1E0G
 
 | | LYSM Domain from E.coli MLTD | | Descriptor: | Membrane-bound lytic murein transglycosylase D | | Authors: | Bateman, A, Bycroft, M. | | Deposit date: | 2000-03-27 | | Release date: | 2000-06-21 | | Last modified: | 2019-09-25 | | Method: | SOLUTION NMR | | Cite: | The Structure of a Lysm Domain from E.Coli Membrane Bound Lytic Murein Transglycosylase D (Mltd)
J.Mol.Biol., 299, 2000
|
|
6XWE
 
 | | Crystal structure of LYK3 ectodomain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ACETONITRILE, LysM domain receptor-like kinase 3, ... | | Authors: | Gysel, K, Blaise, M, Andersen, K.R. | | Deposit date: | 2020-01-23 | | Release date: | 2020-08-12 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Ligand-recognizing motifs in plant LysM receptors are major determinants of specificity.
Science, 369, 2020
|
|
6Q40
 
 | | A secreted LysM effector of the wheat pathogen Zymoseptoria tritici protects the fungal hyphae against chitinase hydrolysis through ligand-dependent polymerisation of LysM homodimers | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, LysM domain-containing protein | | Authors: | Mesters, J.R, Saleem-Batcha, R, Sanchez-Vallet, A, Thomma, B.P.H.J. | | Deposit date: | 2018-12-05 | | Release date: | 2019-10-16 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.412 Å) | | Cite: | A secreted LysM effector protects fungal hyphae through chitin-dependent homodimer polymerization.
Plos Pathog., 16, 2020
|
|
2DJP
 
 | | The solution structure of the LysM domain of human hypothetical protein SB145 | | Descriptor: | Hypothetical protein SB145 | | Authors: | Sasagawa, A, Tochio, N, Saito, K, Koshiba, S, Inoue, M, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2006-04-05 | | Release date: | 2006-10-05 | | Last modified: | 2022-03-09 | | Method: | SOLUTION NMR | | Cite: | The solution structure of the LysM domain of human hypothetical protein SB145
To be Published
|
|
4PXV
 
 | |
4UZ3
 
 | | Crystal structure of the N-terminal LysM domains from the putative NlpC/P60 D,L endopeptidase from T. thermophilus bound to N-acetyl-chitohexaose | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, 2-acetamido-2-deoxy-alpha-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-alpha-D-glucopyranose, ... | | Authors: | Wong, J.E.M.M, Blaise, M. | | Deposit date: | 2014-09-04 | | Release date: | 2015-01-14 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | An Intermolecular Binding Mechanism Involving Multiple Lysm Domains Mediates Carbohydrate Recognition by an Endopeptidase.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4UZ2
 
 | |
5YLG
 
 | | Crystal Structure of LysM domain from pteris ryukyuensis chitinase A | | Descriptor: | 1,2-ETHANEDIOL, Chitinase A, ZINC ION | | Authors: | Ohnuma, T, Kitaoku, Y, Umemoto, N, Numata, T, Fukamizo, T. | | Deposit date: | 2017-10-17 | | Release date: | 2018-10-24 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Crystal Structure of LysM domain from pteris ryukyuensis chitinase A
To Be Published
|
|
5YZK
 
 | | Solution structure of LysM domain from a chitinase derived from Volvox carteri | | Descriptor: | Chitinase, lysozyme | | Authors: | Kitaoku, Y, Nishimura, S, Fukamizo, T, Ohnuma, T. | | Deposit date: | 2017-12-15 | | Release date: | 2018-12-19 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Structures and chitin-binding properties of two N-terminal lysin motifs (LysMs) found in a chitinase from Volvox carteri.
Glycobiology, 29, 2019
|
|
5YZ6
 
 | | Solution structure of LysM domain from a chitinase derived from Volvox carteri | | Descriptor: | Chitinase, lysozyme | | Authors: | Kitaoku, Y, Nishimura, S, Fukamizo, T, Ohnuma, T. | | Deposit date: | 2017-12-13 | | Release date: | 2018-12-19 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Structures and chitin-binding properties of two N-terminal lysin motifs (LysMs) found in a chitinase from Volvox carteri.
Glycobiology, 29, 2019
|
|
7BAX
 
 | | Crystal structure of LYS11 ectodomain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, LysM type receptor kinase | | Authors: | Laursen, M, Cheng, J, Gysel, K, Blaise, M, Andersen, K.R. | | Deposit date: | 2020-12-16 | | Release date: | 2021-11-10 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Kinetic proofreading of lipochitooligosaccharides determines signal activation of symbiotic plant receptors.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
5JCE
 
 | | Crystal structure of OsCEBiP complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Chitin elicitor-binding protein | | Authors: | Chai, J.J, Liu, S.M, Wang, J.Z. | | Deposit date: | 2016-04-15 | | Release date: | 2016-06-08 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Molecular Mechanism for Fungal Cell Wall Recognition by Rice Chitin Receptor OsCEBiP
Structure, 24, 2016
|
|
5JCD
 
 | | Crystal structure of OsCEBiP | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Chitin elicitor-binding protein | | Authors: | Chai, J.J, Liu, S.M, Wang, J.Z. | | Deposit date: | 2016-04-15 | | Release date: | 2017-02-22 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Molecular Mechanism for Fungal Cell Wall Recognition by Rice Chitin Receptor OsCEBiP
Structure, 24, 2016
|
|
5K2L
 
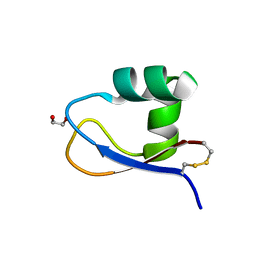 | | Crystal structure of LysM domain from Volvox carteri chitinase | | Descriptor: | 1,2-ETHANEDIOL, Chitinase, lysozyme | | Authors: | Kitaoku, Y, Numata, T, Fukamizo, T, Ohnuma, T. | | Deposit date: | 2016-05-19 | | Release date: | 2017-02-22 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Chitin oligosaccharide binding to the lysin motif of a novel type of chitinase from the multicellular green alga, Volvox carteri.
Plant Mol. Biol., 93, 2017
|
|
7R1L
 
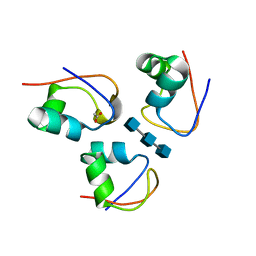 | | Clostridium thermocellum CtCBM50 structure in complex with beta-1,4-GlcNAc trisaccharide | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, Spore coat assembly protein SafA | | Authors: | Ribeiro, D.O, Costa, R, Palma, A.S, Carvalho, A.L. | | Deposit date: | 2022-02-03 | | Release date: | 2023-02-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Unravelling Clostridium thermocellum LysM domains: a molecular view on the cooperative recognition of chitin and peptidoglycan
To Be Published
|
|
5BUM
 
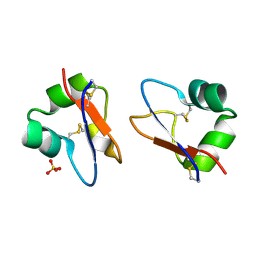 | | Crystal Structure of LysM domain from Equisetum arvense chitinase A | | Descriptor: | Chitinase A, SULFATE ION | | Authors: | Kitaoku, Y, Numata, T, Ohnuma, T, Taira, T, Fukamizo, T. | | Deposit date: | 2015-06-04 | | Release date: | 2016-06-08 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure, mechanism, and phylogeny of LysM-chitinase conjugates specifically found in fern plants.
Plant Sci., 321, 2022
|
|
2MKX
 
 | |
2MPW
 
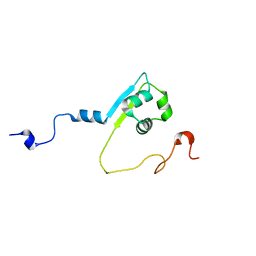 | | Solution structure of the LysM region of the E. coli Intimin periplasmic domain | | Descriptor: | Intimin | | Authors: | Coles, M, Chaubey, M, Leo, J.C, Linke, D, Schuetz, M.C, Goetz, F, Autenrieth, I.B. | | Deposit date: | 2014-06-05 | | Release date: | 2014-11-12 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | The Intimin periplasmic domain mediates dimerisation and binding to peptidoglycan.
Mol.Microbiol., 95, 2015
|
|
4B8V
 
 | | Cladosporium fulvum LysM effector Ecp6 in complex with a beta-1,4- linked N-acetyl-D-glucosamine tetramer | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-alpha-D-glucopyranose, ... | | Authors: | Saleem-Batcha, R, Sanchez-Vallet, A, Hansen, G, Thomma, B.P.H.J, Mesters, J.R. | | Deposit date: | 2012-08-30 | | Release date: | 2013-07-17 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Fungal Effector Ecp6 Outcompetes Host Immune Receptor for Chitin Binding Through Intrachain Lysm Dimerization
Elife, 2, 2013
|
|
4B9H
 
 | | Cladosporium fulvum LysM effector Ecp6 in complex with a beta-1,4- linked N-acetyl-D-glucosamine tetramer: I3C heavy atom derivative | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-alpha-D-glucopyranose, ... | | Authors: | Saleem-Batcha, R, Sanchez-Vallet, A, Hansen, G, Thomma, B.P.H.J, Mesters, J.R. | | Deposit date: | 2012-09-04 | | Release date: | 2013-07-17 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Fungal Effector Ecp6 Outcompetes Host Immune Receptor for Chitin Binding Through Intrachain Lysm Dimerization
Elife, 2, 2013
|
|
4EBZ
 
 | | Crystal structure of the ectodomain of a receptor like kinase | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Chitin elicitor receptor kinase 1, ... | | Authors: | Chai, J, Liu, T, Han, Z, She, J, Wang, J. | | Deposit date: | 2012-03-26 | | Release date: | 2012-06-27 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.792 Å) | | Cite: | Chitin-induced dimerization activates a plant immune receptor.
Science, 336, 2012
|
|
4EBY
 
 | | Crystal structure of the ectodomain of a receptor like kinase | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Chitin elicitor receptor kinase 1, ... | | Authors: | Chai, J, Liu, T, Han, Z, She, J, Wang, J. | | Deposit date: | 2012-03-25 | | Release date: | 2012-06-27 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Chitin-induced dimerization activates a plant immune receptor.
Science, 336, 2012
|
|
1Y7M
 
 | | Crystal Structure of the B. subtilis YkuD protein at 2 A resolution | | Descriptor: | CADMIUM ION, SULFATE ION, hypothetical protein BSU14040 | | Authors: | Bielnicki, J.A, Devedjiev, Y, Derewenda, U, Dauter, Z, Joachimiak, A, Derewenda, Z.S, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2004-12-09 | | Release date: | 2005-03-01 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | B. subtilis ykuD protein at 2.0 A resolution: insights into the structure and function of a novel, ubiquitous family of bacterial enzymes.
Proteins, 62, 2006
|
|
3ZQD
 
 | | B. subtilis L,D-transpeptidase | | Descriptor: | L, D-TRANSPEPTIDASE YKUD | | Authors: | Lecoq, L, Simorre, J.-P, Bougault, C, Arthur, M, Hugonnet, J.-E, Veckerle, C, Pessey, O. | | Deposit date: | 2011-06-09 | | Release date: | 2012-05-23 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Dynamics Induced by Beta-Lactam Antibiotics in the Active Site of Bacillus subtilis L,D-Transpeptidase.
Structure, 20, 2012
|
|
4A1J
 
 | | Ykud L,D-transpeptidase from B.subtilis | | Descriptor: | PUTATIVE L, D-TRANSPEPTIDASE YKUD, SULFATE ION | | Authors: | Blaise, M, Fuglsang Midtgaard, S, Roi Midtgaard, S, Boesen, T, Thirup, S. | | Deposit date: | 2011-09-15 | | Release date: | 2012-09-26 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of Ykud
To be Published
|
|
