8W5J
 
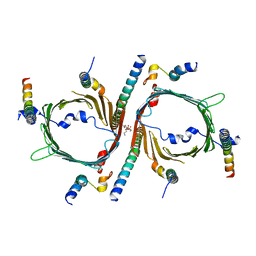 | | Cryo-EM structure of the yeast TOM core complex (from TOM-TIM23 complex) | | Descriptor: | (2R)-3-{[(S)-(2-aminoethoxy)(hydroxy)phosphoryl]oxy}-2-(tetradecanoyloxy)propyl tetradecanoate, Mitochondrial import receptor subunit TOM22, Mitochondrial import receptor subunit TOM40, ... | | Authors: | Wang, Q, Guan, Z.Y, Zhuang, J.J, Huang, R, Yin, P. | | Deposit date: | 2023-08-27 | | Release date: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | The architecture of substrate-engaged TOM-TIM23 supercomplex reveals preprotein proximity sites for mitochondrial protein translocation.
Cell Discov, 10, 2024
|
|
8W5K
 
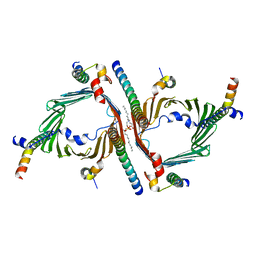 | | Cryo-EM structure of the yeast TOM core complex crosslinked by BS3 (from TOM-TIM23 complex) | | Descriptor: | (2R)-3-{[(S)-(2-aminoethoxy)(hydroxy)phosphoryl]oxy}-2-(tetradecanoyloxy)propyl tetradecanoate, Mitochondrial import receptor subunit TOM22, Mitochondrial import receptor subunit TOM40, ... | | Authors: | Wang, Q, Guan, Z.Y, Zhuang, J.J, Huang, R, Yin, P. | | Deposit date: | 2023-08-27 | | Release date: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | The architecture of substrate-engaged TOM-TIM23 supercomplex reveals preprotein proximity sites for mitochondrial protein translocation.
Cell Discov, 10, 2024
|
|
8J0O
 
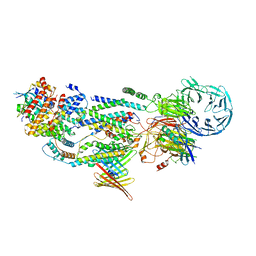 | | cryo-EM structure of human EMC and VDAC | | Descriptor: | ER membrane protein complex subunit 1, ER membrane protein complex subunit 10, ER membrane protein complex subunit 2, ... | | Authors: | Li, M, Zhang, C, Wu, J, Lei, M. | | Deposit date: | 2023-04-11 | | Release date: | 2024-04-03 | | Last modified: | 2024-04-17 | | Method: | ELECTRON MICROSCOPY (3.32 Å) | | Cite: | Structural insights into human EMC and its interaction with VDAC.
Aging (Albany NY), 16, 2024
|
|
8HCO
 
 | | Substrate-engaged TOM complex from yeast | | Descriptor: | Mitochondrial import receptor subunit TOM22, Mitochondrial import receptor subunit TOM40, Mitochondrial import receptor subunit TOM5, ... | | Authors: | Zhou, X.Y, Yang, Y.Q, Wang, G.P, Wang, S.S. | | Deposit date: | 2022-11-02 | | Release date: | 2023-09-13 | | Last modified: | 2023-12-27 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Molecular pathway of mitochondrial preprotein import through the TOM-TIM23 supercomplex.
Nat.Struct.Mol.Biol., 30, 2023
|
|
8B4I
 
 | | Cryo-EM structure of the Neurospora crassa TOM core complex at 3.3 angstrom | | Descriptor: | 2-[2-[(1~{S},2~{S},4~{S},5'~{R},6~{R},7~{S},8~{R},9~{S},12~{S},13~{R},16~{S})-5',7,9,13-tetramethylspiro[5-oxapentacyclo[10.8.0.0^{2,9}.0^{4,8}.0^{13,18}]icos-18-ene-6,2'-oxane]-16-yl]oxyethyl]propane-1,3-diol, DIUNDECYL PHOSPHATIDYL CHOLINE, Mitochondrial import receptor subunit Tom22, ... | | Authors: | Ornelas, P, Kuehlbrandt, W. | | Deposit date: | 2022-09-20 | | Release date: | 2023-08-09 | | Last modified: | 2023-08-30 | | Method: | ELECTRON MICROSCOPY (3.32 Å) | | Cite: | Two conformations of the Tom20 preprotein receptor in the TOM holo complex.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
7TCV
 
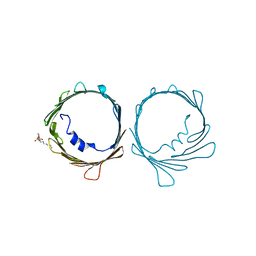 | | VDAC K12E mutant | | Descriptor: | 1,2-DIMYRISTOYL-RAC-GLYCERO-3-PHOSPHOCHOLINE, Voltage-dependent anion-selective channel protein 1 | | Authors: | Khan, F, Abramson, J. | | Deposit date: | 2021-12-28 | | Release date: | 2023-01-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.604 Å) | | Cite: | Dynamical control of the mitochondrial beta-barrel channel VDAC by electrostatic and mechanical coupling
To Be Published
|
|
7QI2
 
 | |
7VKU
 
 | | Cryo-EM structure of SAM-Tom40 intermediate complex | | Descriptor: | Mitochondrial import receptor subunit TOM40, Sorting assembly machinery 35 kDa subunit, Sorting assembly machinery 37 kDa subunit, ... | | Authors: | Takeda, H, Tsutsumi, A, Kikkawa, M, Endo, T. | | Deposit date: | 2021-10-01 | | Release date: | 2023-01-04 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Cryo-EM structure of SAM-Tom40 intermediate complex
To Be Published
|
|
7VDD
 
 | | Human TOM complex with cross-linking | | Descriptor: | Mitochondrial import receptor subunit TOM22 homolog, Mitochondrial import receptor subunit TOM40 homolog, Mitochondrial import receptor subunit TOM5 homolog, ... | | Authors: | Liu, D.S, Sui, S.F. | | Deposit date: | 2021-09-06 | | Release date: | 2022-07-13 | | Method: | ELECTRON MICROSCOPY (3.74 Å) | | Cite: | Structural basis of Tom20 and Tom22 cytosolic domains as the human TOM complex receptors.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7VD2
 
 | | Human TOM complex without cross-linking | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, Mitochondrial import receptor subunit TOM22 homolog, Mitochondrial import receptor subunit TOM40 homolog, ... | | Authors: | Liu, D.S, Sui, S.F. | | Deposit date: | 2021-09-06 | | Release date: | 2022-07-13 | | Method: | ELECTRON MICROSCOPY (2.53 Å) | | Cite: | Structural basis of Tom20 and Tom22 cytosolic domains as the human TOM complex receptors.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7VBY
 
 | | Tom core complex with Tom20 and Tom22 subunits. | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, Mitochondrial import receptor subunit TOM22 homolog, Mitochondrial import receptor subunit TOM5 homolog, ... | | Authors: | Liu, D.S, Sui, S.F. | | Deposit date: | 2021-09-01 | | Release date: | 2022-09-07 | | Method: | ELECTRON MICROSCOPY (2.54 Å) | | Cite: | Tom core complex with Tom20 and Tom22 subunits.
To Be Published
|
|
7VC4
 
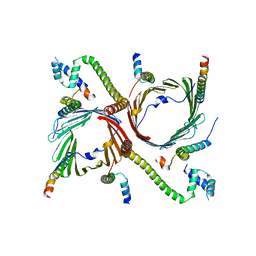 | | Tom complex with Tom22 and Tom20 subunits | | Descriptor: | Mitochondrial import receptor subunit TOM22 homolog, Mitochondrial import receptor subunit TOM40 homolog, Mitochondrial import receptor subunit TOM5 homolog, ... | | Authors: | Liu, D.S, Sui, S.F. | | Deposit date: | 2021-09-01 | | Release date: | 2022-09-07 | | Method: | ELECTRON MICROSCOPY (3.74 Å) | | Cite: | Tom complex with Tom22 and Tom20 subunits
To Be Published
|
|
7E4H
 
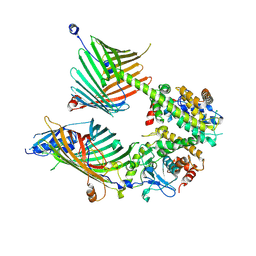 | | Cryo-EM structure of the yeast mitochondrial SAM-Tom40 complex at 3.0 angstrom | | Descriptor: | Mitochondrial import receptor subunit TOM40, Sorting assembly machinery 35 kDa subunit, Sorting assembly machinery 37 kDa subunit, ... | | Authors: | Wang, Q, Guan, Z.Y, Qi, L.B, Yan, C.Y, Yin, P. | | Deposit date: | 2021-02-13 | | Release date: | 2021-09-01 | | Last modified: | 2022-02-16 | | Method: | ELECTRON MICROSCOPY (3.01 Å) | | Cite: | Structural insight into the SAM-mediated assembly of the mitochondrial TOM core complex.
Science, 373, 2021
|
|
7E4I
 
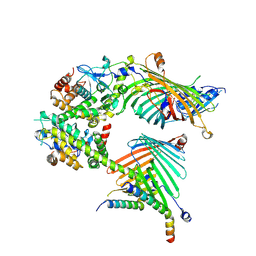 | | Cryo-EM structure of the yeast mitochondrial SAM-Tom40/Tom5/Tom6 complex at 3.0 angstrom | | Descriptor: | Mitochondrial import receptor subunit TOM40, Mitochondrial import receptor subunit TOM5, Mitochondrial import receptor subunit TOM6, ... | | Authors: | Wang, Q, Guan, Z.Y, Qi, L.B, Yan, C.Y, Yin, P. | | Deposit date: | 2021-02-13 | | Release date: | 2021-09-01 | | Last modified: | 2022-02-16 | | Method: | ELECTRON MICROSCOPY (3.05 Å) | | Cite: | Structural insight into the SAM-mediated assembly of the mitochondrial TOM core complex.
Science, 373, 2021
|
|
7NIE
 
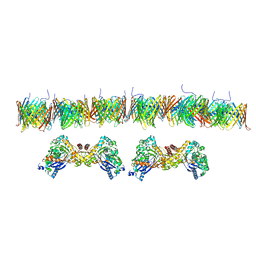 | |
7KUH
 
 | | MicroED structure of mVDAC | | Descriptor: | Voltage-dependent anion-selective channel protein 1 | | Authors: | Martynowycz, M.W, Khan, F, Hattne, J, Abramson, J, Gonen, T. | | Deposit date: | 2020-11-25 | | Release date: | 2020-12-23 | | Last modified: | 2024-03-06 | | Method: | ELECTRON CRYSTALLOGRAPHY (3.12 Å) | | Cite: | MicroED structure of lipid-embedded mammalian mitochondrial voltage-dependent anion channel.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
7CP9
 
 | | Cryo-EM structure of human mitochondrial translocase TOM complex at 3.0 angstrom. | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, Mitochondrial import receptor subunit TOM22 homolog, Mitochondrial import receptor subunit TOM40 homolog, ... | | Authors: | Guan, Z, Yan, L, Wang, Q, Yan, C, Yin, P. | | Deposit date: | 2020-08-06 | | Release date: | 2021-04-28 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural insights into assembly of human mitochondrial translocase TOM complex.
Cell Discov, 7, 2021
|
|
7CK6
 
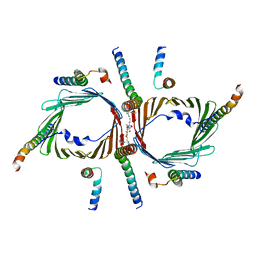 | | Protein translocase of mitochondria | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, Mitochondrial import receptor subunit TOM22 homolog, Mitochondrial import receptor subunit TOM40 homolog, ... | | Authors: | Yang, M, Wang, W, Zhang, L, Chen, X. | | Deposit date: | 2020-07-15 | | Release date: | 2020-11-04 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Atomic structure of human TOM core complex.
Cell Discov, 6, 2020
|
|
6TIQ
 
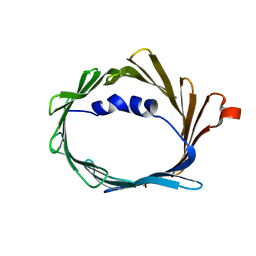 | |
6TIR
 
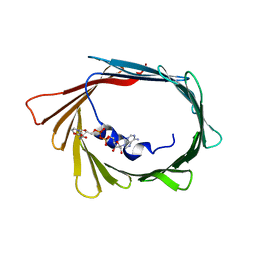 | |
6UCU
 
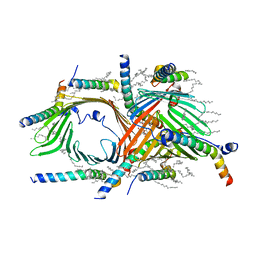 | | Cryo-EM structure of the mitochondrial TOM complex from yeast (dimer) | | Descriptor: | DODECYL-BETA-D-MALTOSIDE, Mitochondrial import receptor subunit TOM22, Mitochondrial import receptor subunit TOM40, ... | | Authors: | Park, E, Tucker, K. | | Deposit date: | 2019-09-17 | | Release date: | 2019-11-06 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.06 Å) | | Cite: | Cryo-EM structure of the mitochondrial protein-import channel TOM complex at near-atomic resolution.
Nat.Struct.Mol.Biol., 26, 2019
|
|
6UCV
 
 | | Cryo-EM structure of the mitochondrial TOM complex from yeast (tetramer) | | Descriptor: | 1,2-DIMYRISTOYL-RAC-GLYCERO-3-PHOSPHOCHOLINE, Mitochondrial import receptor subunit TOM22, Mitochondrial import receptor subunit TOM40, ... | | Authors: | Park, E, Tucker, K. | | Deposit date: | 2019-09-17 | | Release date: | 2019-11-06 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Cryo-EM structure of the mitochondrial protein-import channel TOM complex at near-atomic resolution.
Nat.Struct.Mol.Biol., 26, 2019
|
|
6JNF
 
 | | Cryo-EM structure of the translocator of the outer mitochondrial membrane | | Descriptor: | (2R)-3-{[(S)-(2-aminoethoxy)(hydroxy)phosphoryl]oxy}-2-(tetradecanoyloxy)propyl tetradecanoate, Mitochondrial import receptor subunit TOM22, Mitochondrial import receptor subunit TOM40, ... | | Authors: | Araiso, Y, Tsutsumi, A, Suzuki, J, Yunoki, K, Kawano, S, Kikkawa, M, Endo, T. | | Deposit date: | 2019-03-14 | | Release date: | 2019-10-16 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.81 Å) | | Cite: | Structure of the mitochondrial import gate reveals distinct preprotein paths.
Nature, 575, 2019
|
|
6G73
 
 | | The dynamic nature of the VDAC1 channels in bilayers: human VDAC1 at 3.3 Angstrom resolution | | Descriptor: | 1,2-DIMYRISTOYL-RAC-GLYCERO-3-PHOSPHOCHOLINE, Voltage-dependent anion-selective channel protein 1 | | Authors: | Razeto, A, Gribbon, P, Loew, C. | | Deposit date: | 2018-04-04 | | Release date: | 2019-04-24 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.27 Å) | | Cite: | The dynamic nature of the VDAC1 channels in bilayers as revealed by two crystal structures of the human isoform in bicelles at 2.7 and 3.3 Angstrom resolution: implications for VDAC1 voltage-dependent mechanism and for its oligomerization
To Be Published
|
|
6G6U
 
 | | The dynamic nature of the VDAC1 channels in bilayers: human VDAC1 at 2.7 Angstrom resolution | | Descriptor: | 1,2-DIMYRISTOYL-RAC-GLYCERO-3-PHOSPHOCHOLINE, NITRATE ION, Voltage-dependent anion-selective channel protein 1 | | Authors: | Razeto, A, Gribbon, P, Loew, C. | | Deposit date: | 2018-04-03 | | Release date: | 2019-04-10 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.74 Å) | | Cite: | The dynamic nature of the VDAC1 channels in bilayers as revealed by two crystal structures of the human isoform in bicelles at 2.7 and 3.3 Angstrom resolution: implications for VDAC1 voltage-dependent mechanism and for its oligomerization
To Be Published
|
|
