1M4Z
 
 | | Crystal structure of the N-terminal BAH domain of Orc1p | | Descriptor: | MANGANESE (II) ION, ORIGIN RECOGNITION COMPLEX SUBUNIT 1 | | Authors: | Zhang, Z, Hayashi, M.K, Merkel, O, Stillman, B, Xu, R.-M. | | Deposit date: | 2002-07-05 | | Release date: | 2002-09-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure and function of the BAH-containing domain of Orc1p in epigenetic silencing.
EMBO J., 21, 2002
|
|
1W4S
 
 | |
1ZBX
 
 | | Crystal structure of a Orc1p-Sir1p complex | | Descriptor: | Origin recognition complex subunit 1, Regulatory protein SIR1 | | Authors: | Hsu, H.C, Stillman, B, Xu, R.M. | | Deposit date: | 2005-04-09 | | Release date: | 2005-06-21 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis for origin recognition complex 1 protein-silence information regulator 1 protein interaction in epigenetic silencing
Proc.Natl.Acad.Sci.USA, 102, 2005
|
|
1ZHI
 
 | | Complex of the S. cerevisiae Orc1 and Sir1 interacting domains | | Descriptor: | Origin recognition complex subunit 1, Regulatory protein SIR1 | | Authors: | Hou, Z, Bernstein, D.A, Fox, C.A, Keck, J.L. | | Deposit date: | 2005-04-25 | | Release date: | 2005-06-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural basis of the Sir1-origin recognition complex interaction in transcriptional silencing.
Proc.Natl.Acad.Sci.Usa, 102, 2005
|
|
2FL7
 
 | | S. cerevisiae Sir3 BAH domain | | Descriptor: | Regulatory protein SIR3 | | Authors: | Keck, J.L, Hou, Z, Daner, J.R, Fox, C.A. | | Deposit date: | 2006-01-05 | | Release date: | 2006-05-09 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure of the Sir3 protein bromo adjacent homology (BAH) domain from S. cerevisiae at 1.95 A resolution.
Protein Sci., 15, 2006
|
|
2FVU
 
 | | Structure of the yeast Sir3 BAH domain | | Descriptor: | Regulatory protein SIR3 | | Authors: | Xu, R.M. | | Deposit date: | 2006-01-31 | | Release date: | 2006-09-05 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure and function of the Saccharomyces cerevisiae Sir3 BAH domain.
Mol.Cell.Biol., 26, 2006
|
|
3PT6
 
 | | Crystal structure of mouse DNMT1(650-1602) in complex with DNA | | Descriptor: | DNA (5'-D(*CP*CP*TP*GP*CP*GP*GP*AP*GP*GP*CP*TP*CP*AP*CP*GP*GP*GP*A)-3'), DNA (5'-D(*TP*CP*CP*CP*GP*TP*GP*AP*GP*CP*CP*TP*CP*CP*GP*CP*AP*GP*G)-3'), DNA (cytosine-5)-methyltransferase 1, ... | | Authors: | Song, J, Patel, D.J. | | Deposit date: | 2010-12-02 | | Release date: | 2010-12-29 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure of DNMT1-DNA complex reveals a role for autoinhibition in maintenance DNA methylation.
Science, 331, 2011
|
|
3PTA
 
 | | Crystal structure of human DNMT1(646-1600) in complex with DNA | | Descriptor: | DNA (5'-D(*CP*CP*TP*GP*CP*GP*GP*AP*GP*GP*CP*TP*CP*AP*CP*GP*GP*GP*A)-3'), DNA (5'-D(*TP*CP*CP*CP*GP*TP*GP*AP*GP*CP*CP*TP*CP*CP*GP*CP*AP*GP*G)-3'), DNA (cytosine-5)-methyltransferase 1, ... | | Authors: | Song, J, Patel, D.J. | | Deposit date: | 2010-12-02 | | Release date: | 2010-12-29 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Structure of DNMT1-DNA complex reveals a role for autoinhibition in maintenance DNA methylation.
Science, 331, 2011
|
|
3PT9
 
 | | Crystal structure of mouse DNMT1(731-1602) in the free state | | Descriptor: | DNA (cytosine-5)-methyltransferase 1, S-ADENOSYL-L-HOMOCYSTEINE, ZINC ION | | Authors: | Song, J, Patel, D.J. | | Deposit date: | 2010-12-02 | | Release date: | 2010-12-29 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of DNMT1-DNA complex reveals a role for autoinhibition in maintenance DNA methylation.
Science, 331, 2011
|
|
3AV4
 
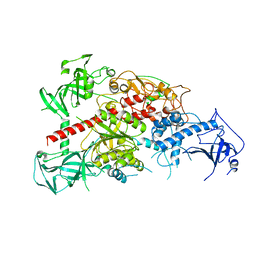 | | Crystal structure of mouse DNA methyltransferase 1 | | Descriptor: | DNA (cytosine-5)-methyltransferase 1, ZINC ION | | Authors: | Takeshita, K, Suetake, I, Yamashita, E, Suga, M, Narita, H, Nakagawa, A, Tajima, S. | | Deposit date: | 2011-02-22 | | Release date: | 2011-05-04 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structural insight into maintenance methylation by mouse DNA methyltransferase 1 (Dnmt1).
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
3AV5
 
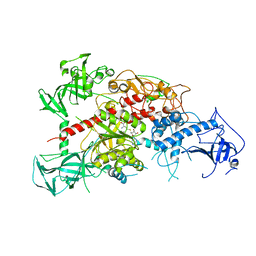 | | Crystal structure of mouse DNA methyltransferase 1 with AdoHcy | | Descriptor: | DNA (cytosine-5)-methyltransferase 1, S-ADENOSYL-L-HOMOCYSTEINE, ZINC ION | | Authors: | Takeshita, K, Suetake, I, Yamashita, E, Suga, M, Narita, H, Nakagawa, A, Tajima, S. | | Deposit date: | 2011-02-22 | | Release date: | 2011-05-04 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | Structural insight into maintenance methylation by mouse DNA methyltransferase 1 (Dnmt1).
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
3AV6
 
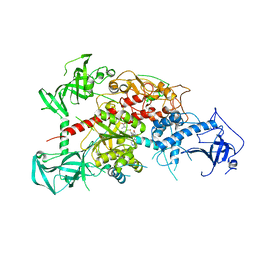 | | Crystal structure of mouse DNA methyltransferase 1 with AdoMet | | Descriptor: | DNA (cytosine-5)-methyltransferase 1, S-ADENOSYLMETHIONINE, ZINC ION | | Authors: | Takeshita, K, Suetake, I, Yamashita, E, Suga, M, Narita, H, Nakagawa, A, Tajima, S. | | Deposit date: | 2011-02-22 | | Release date: | 2011-05-04 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.09 Å) | | Cite: | Structural insight into maintenance methylation by mouse DNA methyltransferase 1 (Dnmt1).
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
3SWR
 
 | |
3TU4
 
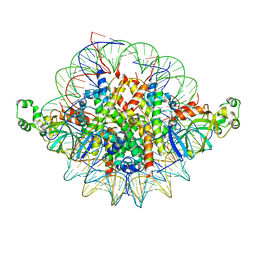 | | Crystal structure of the Sir3 BAH domain in complex with a nucleosome core particle. | | Descriptor: | DNA (146-MER), Histone H2A, Histone H2B 1.1, ... | | Authors: | Armache, K.-J, Garlick, J.D, Canzio, D, Narlikar, G.J, Kingston, R.E. | | Deposit date: | 2011-09-15 | | Release date: | 2011-11-23 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural basis of silencing: Sir3 BAH domain in complex with a nucleosome at 3.0 A resolution.
Science, 334, 2011
|
|
4DA4
 
 | |
4DOV
 
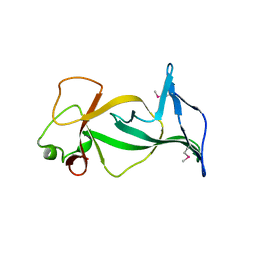 | | Structure of free mouse ORC1 BAH domain | | Descriptor: | Origin recognition complex subunit 1 | | Authors: | Song, J, Patel, D.J. | | Deposit date: | 2012-02-10 | | Release date: | 2012-03-07 | | Last modified: | 2012-04-11 | | Method: | X-RAY DIFFRACTION (1.696 Å) | | Cite: | The BAH domain of ORC1 links H4K20me2 to DNA replication licensing and Meier-Gorlin syndrome.
Nature, 484, 2012
|
|
4DOW
 
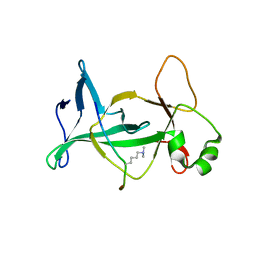 | | Structure of mouse ORC1 BAH domain bound to H4K20me2 | | Descriptor: | Histone H4, Origin recognition complex subunit 1 | | Authors: | Song, J, Patel, D.J. | | Deposit date: | 2012-02-10 | | Release date: | 2012-03-07 | | Last modified: | 2012-04-11 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | The BAH domain of ORC1 links H4K20me2 to DNA replication licensing and Meier-Gorlin syndrome.
Nature, 484, 2012
|
|
4FSX
 
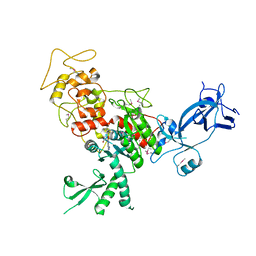 | |
4FT4
 
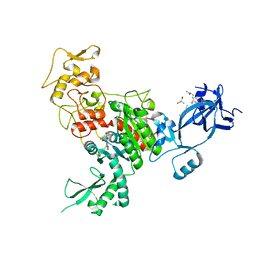 | |
4FT2
 
 | |
4BB7
 
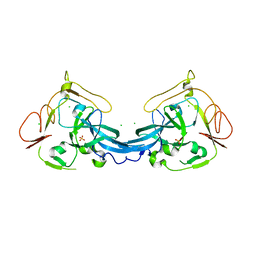 | | Crystal structure of the yeast Rsc2 BAH domain | | Descriptor: | CHLORIDE ION, CHROMATIN STRUCTURE-REMODELING COMPLEX SUBUNIT RSC2, GLYCEROL, ... | | Authors: | Chambers, A.L, Pearl, L.H, Oliver, A.W, Downs, J.A. | | Deposit date: | 2012-09-20 | | Release date: | 2013-08-14 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The Bah Domain of Rsc2 is a Histone H3 Binding Domain.
Nucleic Acids Res., 41, 2013
|
|
4JJN
 
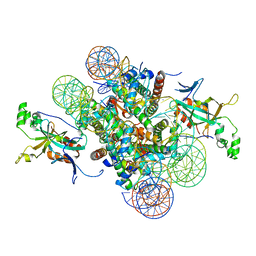 | | Crystal structure of heterochromatin protein Sir3 in complex with a silenced yeast nucleosome | | Descriptor: | DNA (146-MER), Histone H2A.2, Histone H2B.2, ... | | Authors: | Wang, F, Li, G, Mohammed, A, Lu, C, Currie, M, Johnson, A, Moazed, D. | | Deposit date: | 2013-03-08 | | Release date: | 2013-05-15 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.09 Å) | | Cite: | Heterochromatin protein Sir3 induces contacts between the amino terminus of histone H4 and nucleosomal DNA.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4KUL
 
 | | Crystal structure of N-terminal acetylated yeast Sir3 BAH domain V83P mutant | | Descriptor: | Regulatory protein SIR3 | | Authors: | Yang, D, Fang, Q, Wang, M, Ren, R, Wang, H, He, M, Sun, Y, Yang, N, Xu, R.M. | | Deposit date: | 2013-05-22 | | Release date: | 2013-08-07 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.62 Å) | | Cite: | N alpha-acetylated Sir3 stabilizes the conformation of a nucleosome-binding loop in the BAH domain.
Nat.Struct.Mol.Biol., 20, 2013
|
|
4KUI
 
 | | Crystal structure of N-terminal acetylated yeast Sir3 BAH domain | | Descriptor: | ACETIC ACID, ISOPROPYL ALCOHOL, Regulatory protein SIR3 | | Authors: | Yang, D, Fang, Q, Wang, M, Ren, R, Wang, H, He, M, Sun, Y, Yang, N, Xu, R.M. | | Deposit date: | 2013-05-22 | | Release date: | 2013-08-07 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | N alpha-acetylated Sir3 stabilizes the conformation of a nucleosome-binding loop in the BAH domain.
Nat.Struct.Mol.Biol., 20, 2013
|
|
4KUD
 
 | | Crystal structure of N-terminal acetylated Sir3 BAH domain D205N mutant in complex with yeast nucleosome core particle | | Descriptor: | Histone H2A.2, Histone H2B.1, Histone H3, ... | | Authors: | Yang, D, Fang, Q, Wang, M, Ren, R, Wang, H, He, M, Sun, Y, Yang, N, Xu, R.M. | | Deposit date: | 2013-05-22 | | Release date: | 2013-08-07 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.203 Å) | | Cite: | N alpha-acetylated Sir3 stabilizes the conformation of a nucleosome-binding loop in the BAH domain.
Nat.Struct.Mol.Biol., 20, 2013
|
|
