8IOO
 
 | |
7BMF
 
 | |
7BIH
 
 | |
7BJQ
 
 | |
7BHI
 
 | |
7E15
 
 | |
6LRD
 
 | | Structure of RecJ complexed with a 5'-P-dSpacer-modified ssDNA | | Descriptor: | ASP-LEU-PRO-PHE, DNA (5'-D(P*(3DR)P*TP*TP*TP*TP*T)-3'), MANGANESE (II) ION, ... | | Authors: | Cheng, K, Hua, Y. | | Deposit date: | 2020-01-16 | | Release date: | 2020-08-26 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.901335 Å) | | Cite: | Participation of RecJ in the base excision repair pathway of Deinococcus radiodurans.
Nucleic Acids Res., 48, 2020
|
|
6LL8
 
 | | Type II inorganic pyrophosphatase (PPase) from the psychrophilic bacterium Shewanella sp. AS-11, Mg-PNP form | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CALCIUM ION, FLUORIDE ION, ... | | Authors: | Horitani, M, Kusubayashi, K, Oshima, K, Yato, A, Sugimoto, H, Watanabe, K. | | Deposit date: | 2019-12-21 | | Release date: | 2020-03-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | X-ray Crystallography and Electron Paramagnetic Resonance Spectroscopy Reveal Active Site Rearrangement of Cold-Adapted Inorganic Pyrophosphatase.
Sci Rep, 10, 2020
|
|
6LL7
 
 | | Type II inorganic pyrophosphatase (PPase) from the psychrophilic bacterium Shewanella sp. AS-11, Mn-activated form | | Descriptor: | CALCIUM ION, Inorganic pyrophosphatase, MANGANESE (II) ION | | Authors: | Horitani, M, Kusubayashi, K, Oshima, K, Yato, A, Sugimoto, H, Watanabe, K. | | Deposit date: | 2019-12-21 | | Release date: | 2020-03-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | X-ray Crystallography and Electron Paramagnetic Resonance Spectroscopy Reveal Active Site Rearrangement of Cold-Adapted Inorganic Pyrophosphatase.
Sci Rep, 10, 2020
|
|
5X4J
 
 | | The crystal structure of Pyrococcus furiosus RecJ (Zn-soaking) | | Descriptor: | CHLORIDE ION, Uncharacterized protein, ZINC ION | | Authors: | Li, M.J, Yi, G.S, Yu, F, Zhou, H, Chen, J.N, Xu, C.Y, Wang, F.P, Xiao, X, He, J.H, Liu, X.P. | | Deposit date: | 2017-02-13 | | Release date: | 2018-02-14 | | Last modified: | 2019-12-18 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | The crystal structure of Pyrococcus furiosus RecJ implicates it as an ancestor of eukaryotic Cdc45.
Nucleic Acids Res., 45, 2017
|
|
5X4K
 
 | | The complex crystal structure of Pyrococcus furiosus RecJ and CMP | | Descriptor: | CYTIDINE-5'-MONOPHOSPHATE, Uncharacterized protein, ZINC ION | | Authors: | Li, M.J, Yi, G.S, Yu, F, Zhou, H, Chen, J.N, Xu, C.Y, Wang, F.P, Xiao, X, He, J.H, Liu, X.P. | | Deposit date: | 2017-02-13 | | Release date: | 2018-02-14 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.749 Å) | | Cite: | The crystal structure of Pyrococcus furiosus RecJ implicates it as an ancestor of eukaryotic Cdc45.
Nucleic Acids Res., 45, 2017
|
|
5X4H
 
 | | The crystal structure of Pyrococcus furiosus RecJ (wild-type) | | Descriptor: | MAGNESIUM ION, Uncharacterized protein | | Authors: | Li, M.J, Yi, G.S, Yu, F, Zhou, H, Chen, J.N, Xu, C.Y, Wang, F.P, Xiao, X, He, J.H, Liu, X.P. | | Deposit date: | 2017-02-13 | | Release date: | 2018-02-14 | | Last modified: | 2019-12-18 | | Method: | X-RAY DIFFRACTION (2.801 Å) | | Cite: | The crystal structure of Pyrococcus furiosus RecJ implicates it as an ancestor of eukaryotic Cdc45.
Nucleic Acids Res., 45, 2017
|
|
5X4I
 
 | | Pyrococcus furiosus RecJ (D83A, Mn-soaking) | | Descriptor: | CHLORIDE ION, MANGANESE (II) ION, Uncharacterized protein | | Authors: | Li, M.J, Yi, G.S, Yu, F, Zhou, H, Chen, J.N, Xu, C.Y, Wang, F.P, Xiao, X, He, J.H, Liu, X.P. | | Deposit date: | 2017-02-13 | | Release date: | 2018-02-14 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.092 Å) | | Cite: | The crystal structure of Pyrococcus furiosus RecJ implicates it as an ancestor of eukaryotic Cdc45.
Nucleic Acids Res., 45, 2017
|
|
5XSN
 
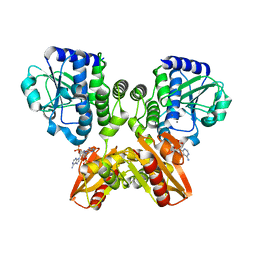 | | The catalytic domain of GdpP with c-di-AMP | | Descriptor: | (2R,3R,3aS,5R,7aR,9R,10R,10aS,12R,14aR)-2,9-bis(6-amino-9H-purin-9-yl)octahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8 ]tetraoxadiphosphacyclododecine-3,5,10,12-tetrol 5,12-dioxide, MANGANESE (II) ION, Phosphodiesterase acting on cyclic dinucleotides | | Authors: | Wang, F, Gu, L. | | Deposit date: | 2017-06-14 | | Release date: | 2018-01-31 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.501 Å) | | Cite: | Structural and biochemical characterization of the catalytic domains of GdpP reveals a unified hydrolysis mechanism for the DHH/DHHA1 phosphodiesterase
Biochem. J., 475, 2018
|
|
5XSI
 
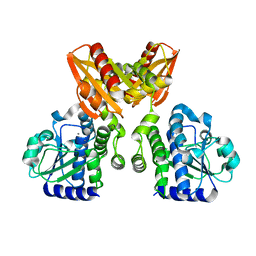 | | The catalytic domain of GdpP | | Descriptor: | MANGANESE (II) ION, Phosphodiesterase acting on cyclic dinucleotides | | Authors: | Wang, F, Gu, L. | | Deposit date: | 2017-06-14 | | Release date: | 2018-01-31 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural and biochemical characterization of the catalytic domains of GdpP reveals a unified hydrolysis mechanism for the DHH/DHHA1 phosphodiesterase
Biochem. J., 475, 2018
|
|
5XSP
 
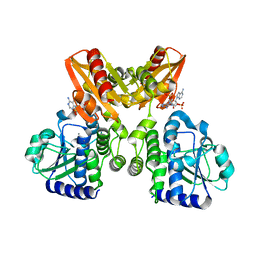 | | The catalytic domain of GdpP with 5'-pApA | | Descriptor: | ADENOSINE-5'-MONOPHOSPHATE, MANGANESE (II) ION, Phosphodiesterase acting on cyclic dinucleotides | | Authors: | Wang, F, Gu, L. | | Deposit date: | 2017-06-15 | | Release date: | 2018-01-31 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.146 Å) | | Cite: | Structural and biochemical characterization of the catalytic domains of GdpP reveals a unified hydrolysis mechanism for the DHH/DHHA1 phosphodiesterase
Biochem. J., 475, 2018
|
|
5XT3
 
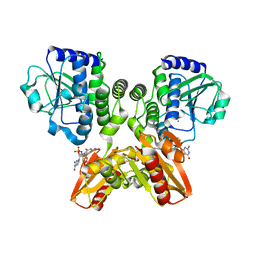 | | The catalytic domain of GdpP with c-di-GMP | | Descriptor: | 9,9'-[(2R,3R,3aS,5S,7aR,9R,10R,10aS,12S,14aR)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-2,9-diyl]bis(2-amino-1,9-dihydro-6H-purin-6-one), MANGANESE (II) ION, Phosphodiesterase acting on cyclic dinucleotides | | Authors: | Wang, F, Gu, L. | | Deposit date: | 2017-06-16 | | Release date: | 2018-01-31 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.591 Å) | | Cite: | Structural and biochemical characterization of the catalytic domains of GdpP reveals a unified hydrolysis mechanism for the DHH/DHHA1 phosphodiesterase
Biochem. J., 475, 2018
|
|
5O25
 
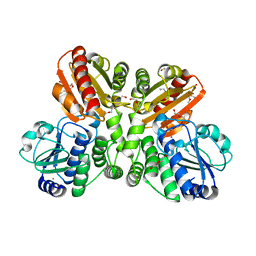 | | Structure of wildtype T.maritima PDE (TM1595) in ligand-free state | | Descriptor: | CALCIUM ION, DI(HYDROXYETHYL)ETHER, MANGANESE (II) ION, ... | | Authors: | Witte, G, Drexler, D, Mueller, M. | | Deposit date: | 2017-05-19 | | Release date: | 2017-10-25 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural and Biophysical Analysis of the Soluble DHH/DHHA1-Type Phosphodiesterase TM1595 from Thermotoga maritima.
Structure, 25, 2017
|
|
5O4Z
 
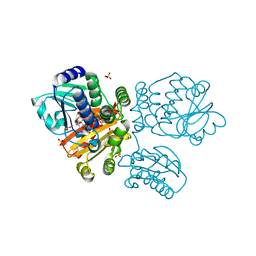 | | Structure of the inactive T.maritima PDE (TM1595) D80N D154N mutant with substrate 5'-pApA | | Descriptor: | ADENOSINE-5'-MONOPHOSPHATE, CHLORIDE ION, DHH/DHHA1-type phosphodiesterase TM1595, ... | | Authors: | Witte, G, Drexler, D, Mueller, M. | | Deposit date: | 2017-05-31 | | Release date: | 2017-10-25 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural and Biophysical Analysis of the Soluble DHH/DHHA1-Type Phosphodiesterase TM1595 from Thermotoga maritima.
Structure, 25, 2017
|
|
5O1U
 
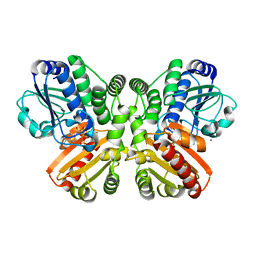 | | Structure of wildtype T.maritima PDE (TM1595) with AMP and Mn2+ | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE MONOPHOSPHATE, CALCIUM ION, ... | | Authors: | Witte, G, Drexler, D, Mueller, M. | | Deposit date: | 2017-05-19 | | Release date: | 2017-10-25 | | Last modified: | 2017-12-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural and Biophysical Analysis of the Soluble DHH/DHHA1-Type Phosphodiesterase TM1595 from Thermotoga maritima.
Structure, 25, 2017
|
|
5O70
 
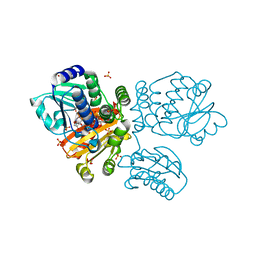 | |
5O58
 
 | | Structure of the inactive T.maritima PDE (TM1595) D80N D154N mutant with substrate 5'-pApG | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, ISOPROPYL ALCOHOL, ... | | Authors: | Witte, G, Drexler, D, Mueller, M. | | Deposit date: | 2017-06-01 | | Release date: | 2017-10-25 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structural and Biophysical Analysis of the Soluble DHH/DHHA1-Type Phosphodiesterase TM1595 from Thermotoga maritima.
Structure, 25, 2017
|
|
5O7F
 
 | | Structure of the inactive T.maritima PDE (TM1595) D80N D154N mutant with GMP and Mn2+ | | Descriptor: | 1,2-ETHANEDIOL, GUANOSINE-5'-MONOPHOSPHATE, MANGANESE (II) ION, ... | | Authors: | Witte, G, Drexler, D, Mueller, M. | | Deposit date: | 2017-06-08 | | Release date: | 2017-10-25 | | Last modified: | 2017-12-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural and Biophysical Analysis of the Soluble DHH/DHHA1-Type Phosphodiesterase TM1595 from Thermotoga maritima.
Structure, 25, 2017
|
|
5IPP
 
 | |
5IZO
 
 | |
