1BJP
 
 | | CRYSTAL STRUCTURE OF 4-OXALOCROTONATE TAUTOMERASE INACTIVATED BY 2-OXO-3-PENTYNOATE AT 2.4 ANGSTROMS RESOLUTION | | Descriptor: | 2-OXO-3-PENTENOIC ACID, 4-OXALOCROTONATE TAUTOMERASE | | Authors: | Taylor, A.B, Czerwinski, R.M, Johnson Junior, W.H, Whitman, C.P, Hackert, M.L. | | Deposit date: | 1998-06-26 | | Release date: | 1998-12-02 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of 4-oxalocrotonate tautomerase inactivated by 2-oxo-3-pentynoate at 2.4 A resolution: analysis and implications for the mechanism of inactivation and catalysis.
Biochemistry, 37, 1998
|
|
7M59
 
 | |
6OGM
 
 | | Crystal structure of apo unFused 4-OT | | Descriptor: | 4-oxalocrotonate tautomerase, GLYCEROL | | Authors: | Medellin, B.P, Whitman, C.P, Zhang, Y.J. | | Deposit date: | 2019-04-03 | | Release date: | 2020-02-26 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.865 Å) | | Cite: | Structural, Kinetic, and Mechanistic Analysis of an Asymmetric 4-Oxalocrotonate Tautomerase Trimer.
Biochemistry, 58, 2019
|
|
7PUO
 
 | | Structure of a fused 4-OT variant engineered for asymmetric Michael addition reactions | | Descriptor: | 2-hydroxymuconate tautomerase,Chains: A,B,C,D,E,F,2-hydroxymuconate tautomerase, CHLORIDE ION, GLYCEROL | | Authors: | Rozeboom, H.J, Thunnissen, A.M.W.H, Poelarends, G.J. | | Deposit date: | 2021-09-30 | | Release date: | 2022-01-26 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Gene Fusion and Directed Evolution to Break Structural Symmetry and Boost Catalysis by an Oligomeric C-C Bond-Forming Enzyme.
Angew.Chem.Int.Ed.Engl., 61, 2022
|
|
7XUY
 
 | |
2FM7
 
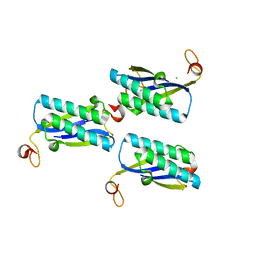 | |
3RY0
 
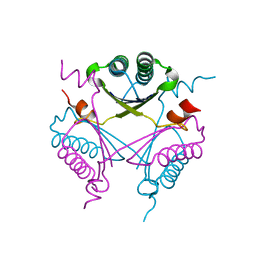 | | Crystal structure of TomN, a 4-Oxalocrotonate Tautomerase homologue in Tomaymycin biosynthetic pathway | | Descriptor: | Putative tautomerase | | Authors: | Zhang, Y, Yan, W.P, Li, W.Z, Whitman, C.P. | | Deposit date: | 2011-05-10 | | Release date: | 2011-08-17 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Kinetic, Crystallographic, and Mechanistic Characterization of TomN: Elucidation of a Function for a 4-Oxalocrotonate Tautomerase Homologue in the Tomaymycin Biosynthetic Pathway.
Biochemistry, 50, 2011
|
|
2X4K
 
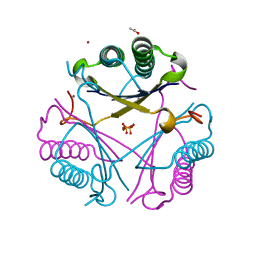 | | Crystal structure of SAR1376, a putative 4-oxalocrotonate tautomerase from the methicillin-resistant Staphylococcus aureus (MRSA) | | Descriptor: | 4-OXALOCROTONATE TAUTOMERASE, ACETATE ION, PHOSPHATE ION, ... | | Authors: | Oke, M, Carter, L.G, Johnson, K.A, Liu, H, Mcmahon, S.A, White, M.F, Naismith, J.H. | | Deposit date: | 2010-02-01 | | Release date: | 2010-07-21 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | The Scottish Structural Proteomics Facility: Targets, Methods and Outputs.
J.Struct.Funct.Genomics, 11, 2010
|
|
4X1C
 
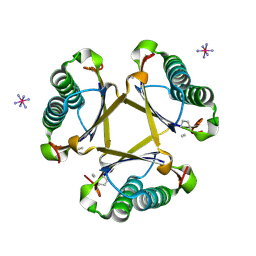 | |
1OTF
 
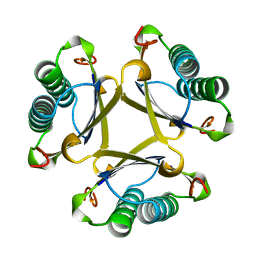 | | 4-OXALOCROTONATE TAUTOMERASE-TRICLINIC CRYSTAL FORM | | Descriptor: | 4-OXALOCROTONATE TAUTOMERASE | | Authors: | Subramanya, H.S, Roper, D.I, Dauter, Z, Dodson, E.J, Davies, G.J, Wilson, K.S, Wigley, D.B. | | Deposit date: | 1995-11-09 | | Release date: | 1996-04-03 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Enzymatic ketonization of 2-hydroxymuconate: specificity and mechanism investigated by the crystal structures of two isomerases.
Biochemistry, 35, 1996
|
|
4X19
 
 | |
6VVR
 
 | |
6FPS
 
 | |
6VVW
 
 | |
6BLM
 
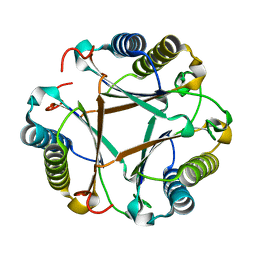 | |
6BGN
 
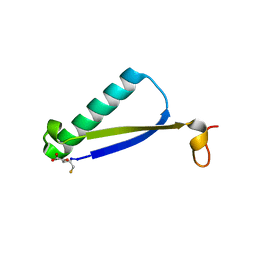 | | Crystal Structure of 4-Oxalocrotonate Tautomerase After Incubation with 5-Fluoro-2-hydroxy-2,4-pentadienoate | | Descriptor: | 2-hydroxymuconate tautomerase, 5-fluoranyl-2-oxidanylidene-pentanoic acid, GLYCEROL, ... | | Authors: | Zhang, Y, Li, W, Stack, T. | | Deposit date: | 2017-10-29 | | Release date: | 2018-02-21 | | Last modified: | 2019-12-25 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Inactivation of 4-Oxalocrotonate Tautomerase by 5-Halo-2-hydroxy-2,4-pentadienoates.
Biochemistry, 57, 2018
|
|
6GHW
 
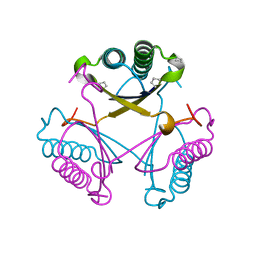 | | Substituting the prolines of 4-oxalocrotonate tautomerase with non-canonical analogue (2S)-3,4-dehydroproline | | Descriptor: | 2-hydroxymuconate tautomerase, CALCIUM ION | | Authors: | Pavkov-Keller, T, Lukesch, M.S, Wiltschi, B, Gruber, K. | | Deposit date: | 2018-05-09 | | Release date: | 2019-03-06 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Substituting the catalytic proline of 4-oxalocrotonate tautomerase with non-canonical analogues reveals a finely tuned catalytic system.
Sci Rep, 9, 2019
|
|
1S0Y
 
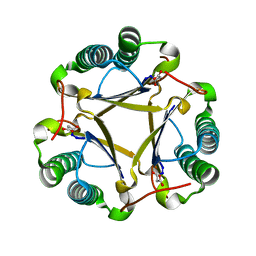 | | The structure of trans-3-chloroacrylic acid dehalogenase, covalently inactivated by the mechanism-based inhibitor 3-bromopropiolate at 2.3 Angstrom resolution | | Descriptor: | MALONIC ACID, alpha-subunit of trans-3-chloroacrylic acid dehalogenase, beta-subunit of trans-3-chloroacrylic acid dehalogenase | | Authors: | de Jong, R.M, Brugman, W, Poelarends, G.J, Whitman, C.P, Dijkstra, B.W. | | Deposit date: | 2004-01-05 | | Release date: | 2004-02-24 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The X-ray structure of trans-3-chloroacrylic acid dehalogenase reveals a novel hydration mechanism in the tautomerase superfamily
J.Biol.Chem., 279, 2004
|
|
5CLO
 
 | |
5CLN
 
 | |
3ABF
 
 | |
4FAZ
 
 | |
4FDX
 
 | |
5TIG
 
 | |
8T9Q
 
 | |
