3IL0
 
 | | The crystal structure of the aminopeptidase P,XAA-pro aminopeptidase from Streptococcus thermophilus | | Descriptor: | Aminopeptidase P; XAA-pro aminopeptidase, GLYCEROL | | Authors: | Zhang, R, Hatzos, C, Cobb, G, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-08-06 | | Release date: | 2009-09-08 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The crystal structure of the aminopeptidase P,XAA-pro aminopeptidase from Streptococcus thermophilus
To be Published
|
|
3I7M
 
 | | N-terminal domain of Xaa-Pro dipeptidase from Lactobacillus brevis. | | Descriptor: | Xaa-Pro dipeptidase | | Authors: | Osipiuk, J, Xu, X, Cui, H, Ng, J, Savchenko, A, Edwards, A.M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-07-08 | | Release date: | 2009-07-14 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | X-ray crystal structure of N-terminal domain of Xaa-Pro dipeptidase from Lactobacillus brevis.
To be Published
|
|
3O5V
 
 | | The Crystal Structure of the Creatinase/Prolidase N-terminal domain of an X-PRO dipeptidase from Streptococcus pyogenes to 1.85A | | Descriptor: | CHLORIDE ION, GLYCEROL, X-PRO dipeptidase | | Authors: | Stein, A.J, Wu, R, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-07-28 | | Release date: | 2010-08-11 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | The Crystal Structure of the Creatinase/Prolidase N-terminal domain of an X-PRO dipeptidase from Streptococcus pyogenes to 1.85A
To be Published
|
|
3OOO
 
 | | The structure of a proline dipeptidase from Streptococcus agalactiae 2603V | | Descriptor: | Proline dipeptidase | | Authors: | Fan, Y, Wu, R, Morales, J, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-08-31 | | Release date: | 2010-09-22 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | The structure of a proline dipeptidase from Streptococcus agalactiae 2603V
To be Published
|
|
3OVK
 
 | |
3PEB
 
 | | The Structure of a Creatine_N Superfamily domain of a dipeptidase from Streptococcus thermophilus. | | Descriptor: | 1,2-ETHANEDIOL, 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, Dipeptidase, ... | | Authors: | Cuff, M.E, Mack, J.C, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-10-25 | | Release date: | 2010-11-03 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | The Structure of a Creatine_N Superfamily domain of a dipeptidase from Streptococcus thermophilus.
TO BE PUBLISHED
|
|
3PN9
 
 | |
3QOC
 
 | | Crystal structure of N-terminal domain (Creatinase/Prolidase like domain) of putative metallopeptidase from Corynebacterium diphtheriae | | Descriptor: | CHLORIDE ION, Putative metallopeptidase, SULFATE ION | | Authors: | Nocek, B, Stein, A, Marshall, N, Putagunta, R, Feldmann, B, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2011-02-09 | | Release date: | 2011-03-23 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystal structure of N-terminal domain (Creatinase/Prolidase like domain) of putative metallopeptidase from Corynebacterium diphtheriae
TO BE PUBLISHED
|
|
5E2C
 
 | | Crystal structure of N-terminal domain of cytoplasmic peptidase PepQ from Mycobacterium tuberculosis H37Rv | | Descriptor: | Xaa-Pro dipeptidase | | Authors: | Chang, C, Endres, L, Endres, M, SACCHETTINI, J, JOACHIMIAK, A, Midwest Center for Structural Genomics (MCSG), Structures of Mtb Proteins Conferring Susceptibility to Known Mtb Inhibitors (MTBI) | | Deposit date: | 2015-09-30 | | Release date: | 2015-10-14 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of N-terminal domain of cytoplasmic peptidase PepQ from Mycobacterium tuberculosis H37Rv
To Be Published
|
|
7N02
 
 | |
1CHM
 
 | | ENZYMATIC MECHANISM OF CREATINE AMIDINOHYDROLASE AS DEDUCED FROM CRYSTAL STRUCTURES | | Descriptor: | CARBAMOYL SARCOSINE, CREATINE AMIDINOHYDROLASE | | Authors: | Hoeffken, H.W, Knof, S.H, Bartlett, P.A, Huber, R, Moellering, H, Schumacher, G. | | Deposit date: | 1993-07-19 | | Release date: | 1994-04-30 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Enzymatic mechanism of creatine amidinohydrolase as deduced from crystal structures.
J.Mol.Biol., 214, 1990
|
|
6YO9
 
 | |
7YTO
 
 | |
4ZNG
 
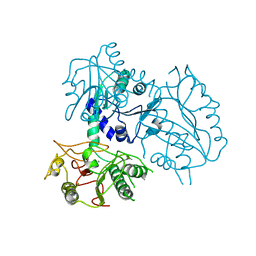 | |
3CTZ
 
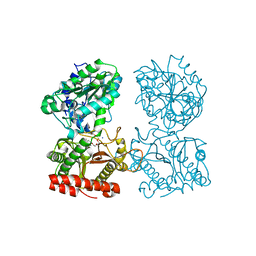 | | Structure of human cytosolic X-prolyl aminopeptidase | | Descriptor: | CALCIUM ION, HEXAETHYLENE GLYCOL, MANGANESE (II) ION, ... | | Authors: | Li, X, Lou, Z, Rao, Z. | | Deposit date: | 2008-04-15 | | Release date: | 2008-05-27 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure of human cytosolic X-prolyl aminopeptidase: a double Mn(II)-dependent dimeric enzyme with a novel three-domain subunit
J.Biol.Chem., 283, 2008
|
|
2HOW
 
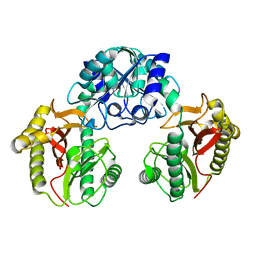 | |
7K3U
 
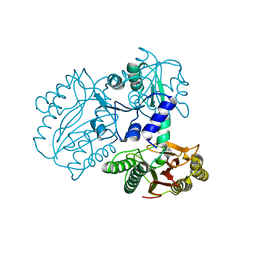 | |
1PV9
 
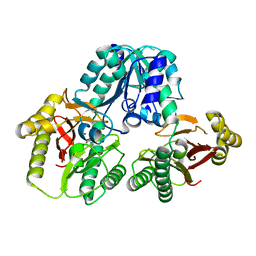 | | Prolidase from Pyrococcus furiosus | | Descriptor: | Xaa-Pro dipeptidase, ZINC ION | | Authors: | Maher, M.J, Ghosh, M, Grunden, A.M, Menon, A.L, Adams, M.W, Freeman, H.C, Guss, J.M. | | Deposit date: | 2003-06-27 | | Release date: | 2004-03-23 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of the Prolidase from Pyrococcus furiosus.
Biochemistry, 43, 2004
|
|
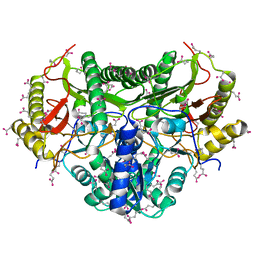
1KP0
 
 | |
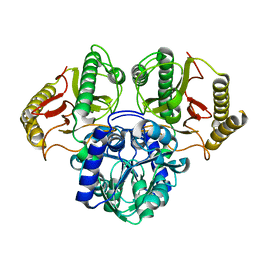
6TWJ
 
 | |
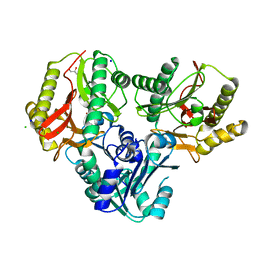
2ZSG
 
 | |
4EGE
 
 | |
6TWK
 
 | | Substrate bound structure of the Ectoine utilization protein EutD (DoeA) from Halomonas elongata | | Descriptor: | (2~{R})-4-azanyl-2-[[(1~{S})-1-oxidanylethyl]amino]butanoic acid, (4S)-2-METHYL-1,4,5,6-TETRAHYDROPYRIMIDINE-4-CARBOXYLIC ACID, Ectoine hydrolase DoeA | | Authors: | Mais, C.-N, Altegoer, F, Bange, G. | | Deposit date: | 2020-01-13 | | Release date: | 2020-05-20 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Degradation of the microbial stress protectants and chemical chaperones ectoine and hydroxyectoine by a bacterial hydrolase-deacetylase complex.
J.Biol.Chem., 295, 2020
|
|
4FKC
 
 | | Recombinant prolidase from Thermococcus sibiricus | | Descriptor: | CADMIUM ION, Xaa-Pro aminopeptidase | | Authors: | Trofimov, A.A, Slutskaya, E.S, Polyakov, K.M, Dorovatovskii, P.V, Gumerov, V.M, Popov, V.O. | | Deposit date: | 2012-06-13 | | Release date: | 2012-11-07 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Influence of intermolecular contacts on the structure of recombinant prolidase from Thermococcus sibiricus.
Acta Crystallogr.,Sect.F, 68, 2012
|
|
6XMR
 
 | |
