6VBG
 
 | | Lactose permease complex with thiodigalactoside and nanobody 9043 | | Descriptor: | Galactoside permease, beta-D-galactopyranose-(1-1)-1-thio-beta-D-galactopyranose, nanobody 9043, ... | | Authors: | Kumar, H, Stroud, R.M, Kaback, H.R, Finer-Moore, J, Smirnova, I, Kasho, V, Pardon, E, Steyart, J. | | Deposit date: | 2019-12-18 | | Release date: | 2020-11-25 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Diversity in kinetics correlated with structure in nano body-stabilized LacY.
Plos One, 15, 2020
|
|
6C9W
 
 | | Crystal Structure of a ligand bound LacY/Nanobody Complex | | Descriptor: | 4-nitrophenyl alpha-D-galactopyranoside, Lactose permease, Nanobody9047, ... | | Authors: | Kumar, H, Finer-Moore, J.S, Jiang, X, Smirnova, I, Kasho, V, Pardon, E, Steyaert, J, Kaback, H.R, Stroud, R.M. | | Deposit date: | 2018-01-29 | | Release date: | 2018-08-15 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal Structure of a ligand-bound LacY-Nanobody Complex.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
5GXB
 
 | | crystal structure of a LacY/Nanobody complex | | Descriptor: | Lactose permease, nanobody | | Authors: | Jiang, X, Wu, J.P, Yan, N, Kaback, H.R. | | Deposit date: | 2016-09-16 | | Release date: | 2016-10-26 | | Last modified: | 2022-10-12 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Crystal structure of a LacY-nanobody complex in a periplasmic-open conformation.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
4ZYR
 
 | | Crystal structure of E. coli Lactose permease G46W/G262W bound to p-nitrophenyl alpha-D-galactopyranoside (alpha-NPG) | | Descriptor: | 4-nitrophenyl alpha-D-galactopyranoside, Lactose permease, nonyl beta-D-glucopyranoside | | Authors: | Kumar, H, Finer-Moore, J.S, Kaback, H.R, Stroud, R.M. | | Deposit date: | 2015-05-22 | | Release date: | 2015-07-29 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.312 Å) | | Cite: | Structure of LacY with an alpha-substituted galactoside: Connecting the binding site to the protonation site.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
2Y5Y
 
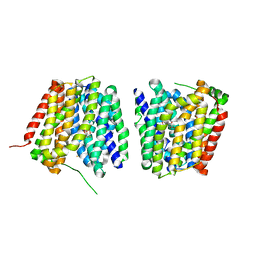 | | Crystal structure of LacY in complex with an affinity inactivator | | Descriptor: | 2-sulfanylethyl beta-D-galactopyranoside, BARIUM ION, LACTOSE PERMEASE | | Authors: | Chaptal, V, Kwon, S, Sawaya, M.R, Guan, L, Kaback, H.R, Abramson, J. | | Deposit date: | 2011-01-19 | | Release date: | 2011-06-15 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.38 Å) | | Cite: | Crystal Structure of Lactose Permease in Complex with an Affinity Inactivator Yields Unique Insight Into Sugar Recognition.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
2V8N
 
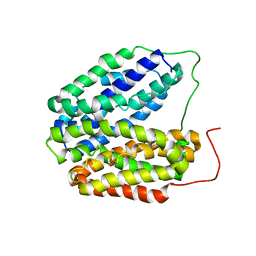 | | Wild-type Structure of Lactose Permease | | Descriptor: | LACTOSE PERMEASE | | Authors: | Guan, L, Mirza, O, Verner, G, Iwata, S, Kaback, H.R. | | Deposit date: | 2007-08-09 | | Release date: | 2007-09-11 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Structural Determination of Wild-Type Lactose Permease.
Proc.Natl.Acad.Sci.USA, 104, 2007
|
|
2CFQ
 
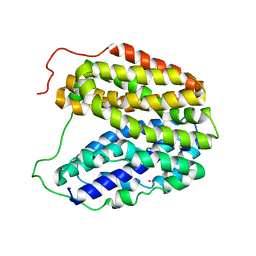 | | Sugar Free Lactose Permease at neutral pH | | Descriptor: | LACTOSE PERMEASE, MERCURY (II) ION | | Authors: | Mirza, O, Guan, L, Verner, G, Iwata, S, Kaback, H.R. | | Deposit date: | 2006-02-22 | | Release date: | 2006-03-13 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Structural Evidence for Induced Fit and a Mechanism for Sugar/H(+) Symport in Lacy.
Embo J., 25, 2006
|
|
2CFP
 
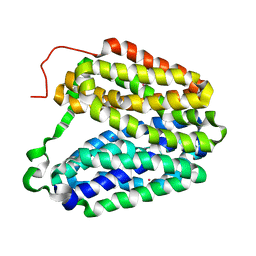 | | Sugar Free Lactose Permease at acidic pH | | Descriptor: | LACTOSE PERMEASE, MERCURY (II) ION | | Authors: | Mirza, O, Guan, L, Verner, G, Iwata, S, Kaback, H.R. | | Deposit date: | 2006-02-22 | | Release date: | 2006-03-13 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structural Evidence for Induced Fit and a Mechanism for Sugar/H(+) Symport in Lacy.
Embo J., 25, 2006
|
|
1PV7
 
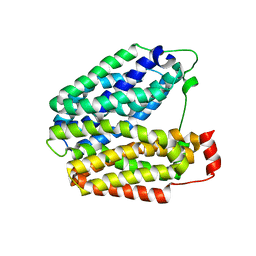 | | Crystal structure of lactose permease with TDG | | Descriptor: | Lactose permease, beta-D-galactopyranose-(1-1)-1-thio-beta-D-galactopyranose | | Authors: | Abramson, J, Smirnova, I, Kasho, V, Verner, G, Kaback, H.R, Iwata, S. | | Deposit date: | 2003-06-26 | | Release date: | 2003-08-12 | | Last modified: | 2021-11-10 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Structure and mechanism of the lactose permease of Escherichia coli
SCIENCE, 301, 2003
|
|
1PV6
 
 | | Crystal structure of lactose permease | | Descriptor: | Lactose permease | | Authors: | Abramson, J, Smirnova, I, Kasho, V, Verner, G, Kaback, H.R, Iwata, S. | | Deposit date: | 2003-06-26 | | Release date: | 2003-08-12 | | Last modified: | 2021-11-10 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structure and mechanism of the lactose permease of Escherichia coli
SCIENCE, 301, 2003
|
|
