8PYV
 
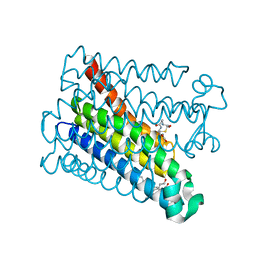 | | Structure of Human PS-1 GSH-analog complex, solved at wavelength 2.755 A | | Descriptor: | L-gamma-glutamyl-S-(2-biphenyl-4-yl-2-oxoethyl)-L-cysteinylglycine, PALMITIC ACID, Prostaglandin E synthase | | Authors: | Duman, R, El Omari, K, Mykhaylyk, V, Orr, C, Wagner, A, Vogeley, L, Brown, D.G. | | Deposit date: | 2023-07-26 | | Release date: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Experimental phasing opportunities for macromolecular crystallography at very long wavelengths.
Commun Chem, 6, 2023
|
|
6VL4
 
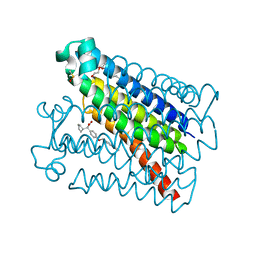 | | Crystal Structure of mPGES-1 bound to DG-031 | | Descriptor: | (2R)-cyclopentyl{4-[(quinolin-2-yl)methoxy]phenyl}acetic acid, Prostaglandin E synthase, TETRAETHYLENE GLYCOL, ... | | Authors: | Ho, J.D, Lee, M.R, Rauch, C.T, Aznavour, K, Park, J.S, Luz, J.G, Antonysamy, S, Condon, B, Maletic, M, Zhang, A, Hickey, M.J, Hughes, N.E, Chandrasekhar, S, Sloan, A.V, Gooding, K, Harvey, A, Yu, X.P, Kahl, S.D, Norman, B.H. | | Deposit date: | 2020-01-22 | | Release date: | 2020-12-02 | | Last modified: | 2020-12-16 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structure-based, multi-targeted drug discovery approach to eicosanoid inhibition: Dual inhibitors of mPGES-1 and 5-lipoxygenase activating protein (FLAP).
Biochim Biophys Acta Gen Subj, 1865, 2020
|
|
6VGI
 
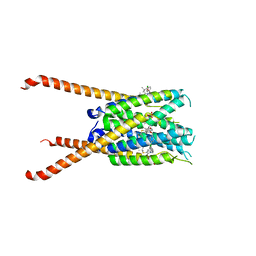 | | Crystal Structures of FLAP bound to MK-866 | | Descriptor: | 3-[3-(tert-butylsulfanyl)-1-[(4-chlorophenyl)methyl]-5-(propan-2-yl)-1H-indol-2-yl]-2,2-dimethylpropanoic acid, 5-lipoxygenase-activating protein, SULFATE ION | | Authors: | Ho, J.D, Lee, M.R, Rauch, C.T, Aznavour, K, Park, J.S, Luz, J.G, Antonysamy, S, Condon, B, Maletic, M, Zhang, A, Hickey, M.J, Hughes, N.E, Chandrasekhar, S, Sloan, A.V, Gooding, K, Harvey, A, Yu, X.P, Kahl, S.D, Norman, B.H. | | Deposit date: | 2020-01-08 | | Release date: | 2020-12-02 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Structure-based, multi-targeted drug discovery approach to eicosanoid inhibition: Dual inhibitors of mPGES-1 and 5-lipoxygenase activating protein (FLAP).
Biochim Biophys Acta Gen Subj, 1865, 2020
|
|
6VGC
 
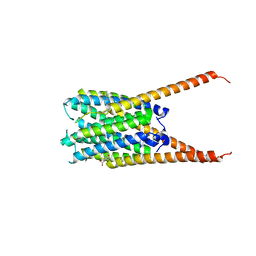 | | Crystal Structures of FLAP bound to DG-031 | | Descriptor: | (2R)-cyclopentyl{4-[(quinolin-2-yl)methoxy]phenyl}acetic acid, 5-lipoxygenase-activating protein, CALCIUM ION, ... | | Authors: | Ho, J.D, Lee, M.R, Rauch, C.T, Aznavour, K, Park, J.S, Luz, J.G, Antonysamy, S, Condon, B, Maletic, M, Zhang, A, Hickey, M.J, Hughes, N.E, Chandrasekhar, S, Sloan, A.V, Gooding, K, Harvey, A, Yu, X.P, Kahl, S.D, Norman, B.H. | | Deposit date: | 2020-01-07 | | Release date: | 2020-12-02 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | Structure-based, multi-targeted drug discovery approach to eicosanoid inhibition: Dual inhibitors of mPGES-1 and 5-lipoxygenase activating protein (FLAP).
Biochim Biophys Acta Gen Subj, 1865, 2020
|
|
6SSW
 
 | |
6SSU
 
 | |
6SSS
 
 | | Crystal structure of Human Microsomal Glutathione S-Transferase 2 | | Descriptor: | 1-(8Z-hexadecenoyl)-sn-glycerol, GLYCEROL, Microsomal glutathione S-transferase 2, ... | | Authors: | Thulasingam, M, Nji, E, Haeggstrom, J.Z. | | Deposit date: | 2019-09-09 | | Release date: | 2021-02-03 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.498 Å) | | Cite: | Crystal structures of human MGST2 reveal synchronized conformational changes regulating catalysis.
Nat Commun, 12, 2021
|
|
6SSR
 
 | |
6R7D
 
 | | Crystal structure of LTC4S in complex with AZ13690257 | | Descriptor: | (1~{S},2~{S})-2-[5-[cyclopropylmethyl(naphthalen-1-yl)amino]-4-methoxy-pyrimidin-2-yl]carbonylcyclopropane-1-carboxylic acid, DODECYL-BETA-D-MALTOSIDE, Leukotriene C4 synthase, ... | | Authors: | Kack, H, Ek, M. | | Deposit date: | 2019-03-28 | | Release date: | 2019-08-28 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Discovery of the Oral Leukotriene C4 Synthase Inhibitor (1S,2S)-2-({5-[(5-Chloro-2,4-difluorophenyl)(2-fluoro-2-methylpropyl)amino]-3-methoxypyrazin-2-yl}carbonyl)cyclopropanecarboxylic Acid (AZD9898) as a New Treatment for Asthma.
J.Med.Chem., 62, 2019
|
|
5TL9
 
 | | crystal structure of mPGES-1 bound to inhibitor | | Descriptor: | 2-{2-[(1S,2S)-2-{[1-(8-methylquinolin-2-yl)piperidine-4-carbonyl]amino}cyclopentyl]ethyl}benzoic acid, DI(HYDROXYETHYL)ETHER, GLUTATHIONE, ... | | Authors: | Luz, J.G, Antonysamy, S, Partridge, K, Fisher, M. | | Deposit date: | 2016-10-10 | | Release date: | 2017-03-01 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Discovery and characterization of [(cyclopentyl)ethyl]benzoic acid inhibitors of microsomal prostaglandin E synthase-1.
Bioorg. Med. Chem. Lett., 27, 2017
|
|
5T37
 
 | | crystal structure of mPGES-1 bound to inhibitor | | Descriptor: | 2-chloro-5-{[(2,2-dimethylpropanoyl)amino]methyl}-N-(1H-imidazol-2-yl)benzamide, GLUTATHIONE, Prostaglandin E synthase, ... | | Authors: | Luz, J.G, Antonysamy, S, Partridge, K, Fisher, M. | | Deposit date: | 2016-08-24 | | Release date: | 2017-03-01 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.761 Å) | | Cite: | Discovery and characterization of [(cyclopentyl)ethyl]benzoic acid inhibitors of microsomal prostaglandin E synthase-1.
Bioorg. Med. Chem. Lett., 27, 2017
|
|
5T36
 
 | | Crystal structure of mPGES-1 bound to inhibitor | | Descriptor: | 4-chloro-2-[({(1S,2S)-2-[(2,2-dimethylpropanoyl)amino]cyclopentyl}methyl)amino]benzoic acid, GLUTATHIONE, Prostaglandin E synthase, ... | | Authors: | Luz, J.G, Antonysamy, S, Partridge, K, Fisher, M. | | Deposit date: | 2016-08-24 | | Release date: | 2017-03-01 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Discovery and characterization of [(cyclopentyl)ethyl]benzoic acid inhibitors of microsomal prostaglandin E synthase-1.
Bioorg. Med. Chem. Lett., 27, 2017
|
|
5K0I
 
 | | mpges1 bound to an inhibitor | | Descriptor: | 1,5-anhydro-2,3,4-trideoxy-3-{[(4S)-3,3-dimethyl-1-(8-methylquinolin-2-yl)piperidine-4-carbonyl]amino}-D-erythro-hexitol, GLUTATHIONE, Prostaglandin E synthase, ... | | Authors: | Luz, J.G, Kuklish, S.L. | | Deposit date: | 2016-05-17 | | Release date: | 2016-09-14 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Characterization of 3,3-dimethyl substituted N-aryl piperidines as potent microsomal prostaglandin E synthase-1 inhibitors.
Bioorg.Med.Chem.Lett., 26, 2016
|
|
5IA9
 
 | | The structure of microsomal glutathione transferase 1 in complex with Meisenheimer complex | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1-(S-GLUTATHIONYL)-2,4,6-TRINITROCYCLOHEXA-2,5-DIENE, Microsomal glutathione S-transferase 1, ... | | Authors: | Kuang, Q, Purhonen, P, Jegerschold, C, Morgenstern, R, Hebert, H. | | Deposit date: | 2016-02-21 | | Release date: | 2017-07-12 | | Last modified: | 2017-08-23 | | Method: | ELECTRON CRYSTALLOGRAPHY (3.5 Å) | | Cite: | Dead-end complex, lipid interactions and catalytic mechanism of microsomal glutathione transferase 1, an electron crystallography and mutagenesis investigation.
Sci Rep, 7, 2017
|
|
5I9K
 
 | | The structure of microsomal glutathione transferase 1 | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, GLUTATHIONE, Microsomal glutathione S-transferase 1, ... | | Authors: | Kuang, Q, Purhonen, P, Jegerschold, C, Morgenstern, R, Hebert, H. | | Deposit date: | 2016-02-20 | | Release date: | 2017-07-12 | | Last modified: | 2017-08-23 | | Method: | ELECTRON CRYSTALLOGRAPHY (3.5 Å) | | Cite: | Dead-end complex, lipid interactions and catalytic mechanism of microsomal glutathione transferase 1, an electron crystallography and mutagenesis investigation.
Sci Rep, 7, 2017
|
|
5HV9
 
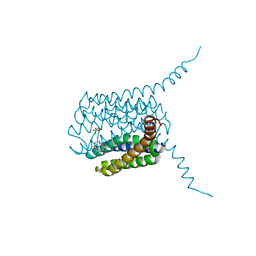 | | Human LTC4S mutant-S36E | | Descriptor: | GLUTATHIONE, Leukotriene C4 synthase, SULFATE ION | | Authors: | Thulasingam, M, Ahmad, H.R.S, Rinaldo-Matthis, A, Haeggstrom, J.Z. | | Deposit date: | 2016-01-28 | | Release date: | 2016-07-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Phosphorylation of Leukotriene C4 Synthase at Serine 36 Impairs Catalytic Activity.
J.Biol.Chem., 291, 2016
|
|
5BQI
 
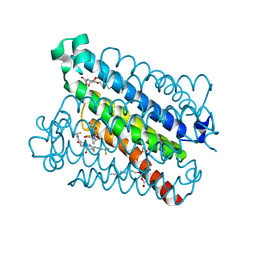 | | Discovery of a Potent and Selective mPGES-1 Inhibitor for the Treatment of Pain | | Descriptor: | (2-hydroxyethoxy)acetaldehyde, 2-(difluoromethyl)-5-{[(2-methylpropanoyl)amino]methyl}-N-{5-methyl-4-[4-(trifluoromethyl)phenyl]-1H-imidazol-2-yl}pyridine-3-carboxamide, GLUTATHIONE, ... | | Authors: | Fisher, M.J, Schiffler, M.A, Kuklish, S.L, Antonysamy, S, Luz, J.G. | | Deposit date: | 2015-05-29 | | Release date: | 2016-04-13 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Discovery and Characterization of 2-Acylaminoimidazole Microsomal Prostaglandin E Synthase-1 Inhibitors.
J.Med.Chem., 59, 2016
|
|
5BQH
 
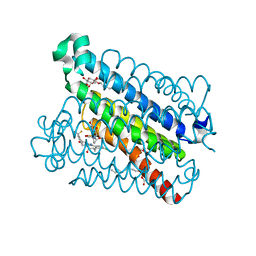 | | Discovery of a Potent and Selective mPGES-1 Inhibitor for the Treatment of Pain | | Descriptor: | (2-hydroxyethoxy)acetaldehyde, GLUTATHIONE, N-[4-(4-chlorophenyl)-1H-imidazol-2-yl]-2-(difluoromethyl)-5-{[(2-methylpropanoyl)amino]methyl}benzamide, ... | | Authors: | Fisher, M.J, Schiffler, M.A, Kuklish, S.L, Antonysamy, S, Luz, J.G. | | Deposit date: | 2015-05-29 | | Release date: | 2016-04-13 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.601 Å) | | Cite: | Discovery and Characterization of 2-Acylaminoimidazole Microsomal Prostaglandin E Synthase-1 Inhibitors.
J.Med.Chem., 59, 2016
|
|
5BQG
 
 | | Crystal Structure of mPGES-1 Bound to an Inhibitor | | Descriptor: | 2-chloro-N-(4-phenyl-1,3-thiazol-2-yl)benzamide, DI(HYDROXYETHYL)ETHER, GLUTATHIONE, ... | | Authors: | Fisher, M.J, Schiffler, M.A, Kuklish, S.L, Antonysamy, S, Luz, J.G. | | Deposit date: | 2015-05-29 | | Release date: | 2016-04-13 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.436 Å) | | Cite: | Discovery and Characterization of 2-Acylaminoimidazole Microsomal Prostaglandin E Synthase-1 Inhibitors.
J.Med.Chem., 59, 2016
|
|
4YL3
 
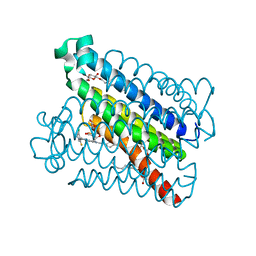 | | Crystal Structures of mPGES-1 Inhibitor Complexes | | Descriptor: | 5-[4-bromo-2-(2-chloro-6-fluorophenyl)-1H-imidazol-5-yl]-2-{[4-(trifluoromethyl)phenyl]ethynyl}pyridine, GLUTATHIONE, Prostaglandin E synthase, ... | | Authors: | Luz, J.G, Antonysamy, S, Kuklish, S.L, Fisher, M.J. | | Deposit date: | 2015-03-04 | | Release date: | 2015-07-08 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.41 Å) | | Cite: | Crystal Structures of mPGES-1 Inhibitor Complexes Form a Basis for the Rational Design of Potent Analgesic and Anti-Inflammatory Therapeutics.
J.Med.Chem., 58, 2015
|
|
4YL1
 
 | | Crystal Structures of mPGES-1 Inhibitor Complexes | | Descriptor: | 5-(4-tert-butylphenyl)-1-[4-(propan-2-yloxy)phenyl]-1H-indole-2-carboxylic acid, DI(HYDROXYETHYL)ETHER, GLUTATHIONE, ... | | Authors: | Luz, J.G, Antonysamy, S, Kuklish, S.L, Fisher, M.J. | | Deposit date: | 2015-03-04 | | Release date: | 2015-06-10 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.41 Å) | | Cite: | Crystal Structures of mPGES-1 Inhibitor Complexes Form a Basis for the Rational Design of Potent Analgesic and Anti-Inflammatory Therapeutics.
J.Med.Chem., 58, 2015
|
|
4YL0
 
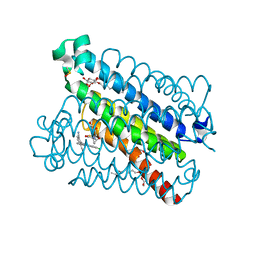 | | Crystal Structures of mPGES-1 Inhibitor Complexes | | Descriptor: | 2-(9-chloro-1H-phenanthro[9,10-d]imidazol-2-yl)benzene-1,3-dicarbonitrile, DI(HYDROXYETHYL)ETHER, GLUTATHIONE, ... | | Authors: | Luz, J.G, Antonysamy, S, Kuklish, S.L, Fisher, M.J. | | Deposit date: | 2015-03-04 | | Release date: | 2015-06-10 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Crystal Structures of mPGES-1 Inhibitor Complexes Form a Basis for the Rational Design of Potent Analgesic and Anti-Inflammatory Therapeutics.
J.Med.Chem., 58, 2015
|
|
4YK5
 
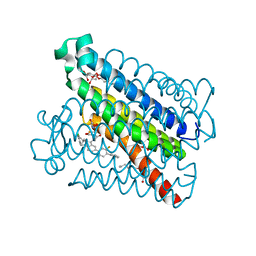 | | Crystal Structures of mPGES-1 Inhibitor Complexes | | Descriptor: | 2-(2-METHOXYETHOXY)ETHANOL, 3-[1-(4-chlorobenzyl)-5-(2-fluoro-2'-methylbiphenyl-4-yl)-3-methyl-1H-indol-2-yl]-2,2-dimethylpropanoic acid, GLUTATHIONE, ... | | Authors: | Luz, J.G, Antonysamy, S, Kuklish, S.L, Fisher, M.J. | | Deposit date: | 2015-03-04 | | Release date: | 2015-07-15 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Crystal Structures of mPGES-1 Inhibitor Complexes Form a Basis for the Rational Design of Potent Analgesic and Anti-Inflammatory Therapeutics.
J.Med.Chem., 58, 2015
|
|
4WAB
 
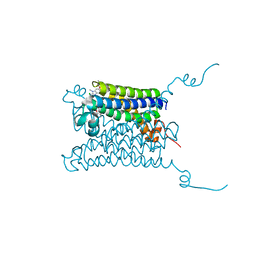 | | Crystal structure of mPGES1 solved by native-SAD phasing | | Descriptor: | 2-[[2,6-bis(chloranyl)-3-[(2,2-dimethylpropanoylamino)methyl]phenyl]amino]-1-methyl-6-(2-methyl-2-oxidanyl-propoxy)-N-[2,2,2-tris(fluoranyl)ethyl]benzimidazole-5-carboxamide, GLUTATHIONE, Prostaglandin E synthase,Leukotriene C4 synthase | | Authors: | Weinert, T, Li, D, Howe, N, Caffrey, M, Wang, M. | | Deposit date: | 2014-08-29 | | Release date: | 2014-12-10 | | Last modified: | 2015-02-11 | | Method: | X-RAY DIFFRACTION (2.704 Å) | | Cite: | Fast native-SAD phasing for routine macromolecular structure determination.
Nat.Methods, 12, 2015
|
|
4NTF
 
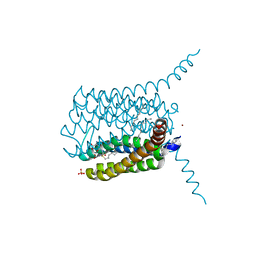 | |
