7C1U
 
 | | Crystal structure of the starter condensation domain of rhizomide synthetase RzmA mutant H140V/R148A in a "product-released" conformation | | Descriptor: | Non-ribosomal peptide synthetase modules | | Authors: | Zhong, L, Diao, X, Zhang, N, Li, F.W, Zhou, H.B, Chen, H.N, Ren, X, Zhang, Y, Wu, D, Bian, X. | | Deposit date: | 2020-05-05 | | Release date: | 2020-11-25 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Engineering and elucidation of the lipoinitiation process in nonribosomal peptide biosynthesis.
Nat Commun, 12, 2021
|
|
2XHG
 
 | | Crystal Structure of the Epimerization Domain from the Initiation Module of Tyrocidine Biosynthesis | | Descriptor: | GLYCEROL, SODIUM ION, TYROCIDINE SYNTHETASE A | | Authors: | Samel, S.-A, Heine, A, Marahiel, M.A, Essen, L.-O. | | Deposit date: | 2010-06-15 | | Release date: | 2011-07-06 | | Last modified: | 2014-10-22 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure of the Epimerization Domain of Tyrocidine Synthetase A
Acta Crystallogr.,Sect.D, 70, 2014
|
|
8F7I
 
 | | The condensation domain of surfactin A synthetase C variant 18b in space group P43212 | | Descriptor: | GLYCEROL, Surfactin synthetase | | Authors: | Frota, N.F, Pistofidis, A, Folger, I.B, Hilvert, D, Schmeing, M. | | Deposit date: | 2022-11-18 | | Release date: | 2023-11-22 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | High-throughput reprogramming of an NRPS condensation domain.
Nat.Chem.Biol., 2024
|
|
5DU9
 
 | | First condensation domain of the calcium-dependent antibiotic synthetase in complex with substrate analogue 2a | | Descriptor: | (2S)-2-amino-N-butyl-propanamide, CDA peptide synthetase I, CHLORIDE ION, ... | | Authors: | Bloudoff, K, Alonzo, D.A, Schmeing, T.M. | | Deposit date: | 2015-09-18 | | Release date: | 2016-03-30 | | Last modified: | 2020-01-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Chemical Probes Allow Structural Insight into the Condensation Reaction of Nonribosomal Peptide Synthetases.
Cell Chem Biol, 23, 2016
|
|
8F7F
 
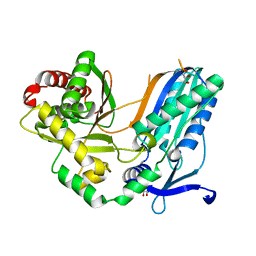 | | The condensation domain of surfactin A synthetase C in space group P43212 | | Descriptor: | GLYCEROL, Surfactin synthetase | | Authors: | Frota, N, Pistofidis, A, Folger, I.B, Hilvert, D, Schmeing, T.M. | | Deposit date: | 2022-11-18 | | Release date: | 2023-11-22 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | High-throughput reprogramming of an NRPS condensation domain.
Nat.Chem.Biol., 2024
|
|
4JN3
 
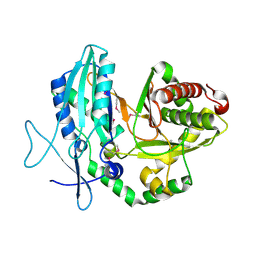 | |
7C1R
 
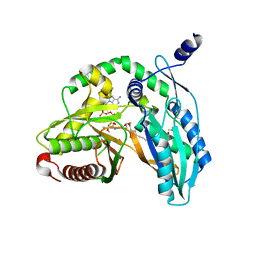 | | Crystal structure of the starter condensation domain of rhizomide synthetase RzmA mutant H140A/R148A in complex with C8-CoA | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Non-ribosomal peptide synthetase modules, OCTANOYL-COENZYME A | | Authors: | Zhong, L, Diao, X, Zhang, N, Li, F.W, Zhou, H.B, Chen, H.N, Ren, X, Zhang, Y, Wu, D, Bian, X. | | Deposit date: | 2020-05-05 | | Release date: | 2020-11-25 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.698 Å) | | Cite: | Engineering and elucidation of the lipoinitiation process in nonribosomal peptide biosynthesis.
Nat Commun, 12, 2021
|
|
5ISW
 
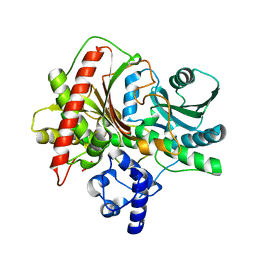 | |
6AD3
 
 | |
5DLK
 
 | | The crystal structure of CT mutant | | Descriptor: | 1,2-ETHANEDIOL, DIMETHYL SULFOXIDE, TqaA | | Authors: | Zhang, J.R, Tang, Y, Zhou, J.H. | | Deposit date: | 2015-09-06 | | Release date: | 2016-09-07 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis of nonribosomal peptide macrocyclization in fungi
Nat.Chem.Biol., 12, 2016
|
|
7C1L
 
 | | Crystal structure of the starter condensation domain of rhizomide synthetase RzmA mutant R148A in complex with C8-CoA | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Non-ribosomal peptide synthetase modules, OCTANOYL-COENZYME A | | Authors: | Zhong, L, Diao, X, Zhang, N, Li, F.W, Zhou, H.B, Chen, H.N, Ren, X, Zhang, Y, Wu, D, Bian, X. | | Deposit date: | 2020-05-05 | | Release date: | 2020-11-25 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Engineering and elucidation of the lipoinitiation process in nonribosomal peptide biosynthesis.
Nat Commun, 12, 2021
|
|
2JGP
 
 | | Structure of the TycC5-6 PCP-C bidomain of the tyrocidine synthetase TycC | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, SODIUM ION, SULFATE ION, ... | | Authors: | Samel, S.A, Schoenafinger, G, Knappe, T.A, Marahiel, M.A, Essen, L.-O. | | Deposit date: | 2007-02-13 | | Release date: | 2007-10-30 | | Last modified: | 2019-07-24 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural and Functional Insights Into a Peptide Bond-Forming Bidomain from a Nonribosomal Peptide Synthetase.
Structure, 15, 2007
|
|
5DUA
 
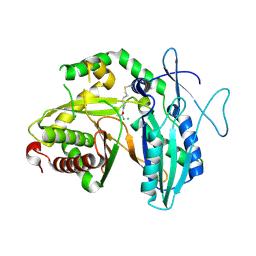 | |
7KW0
 
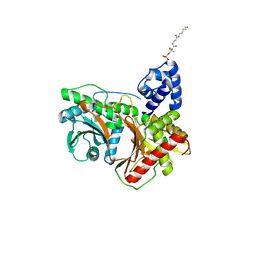 | | Non-ribosomal didomain (stabilised glycine-PCP-C) acceptor bound state | | Descriptor: | N-{2-[(2-aminoethyl)sulfanyl]ethyl}-N~3~-[(2R)-2-hydroxy-3,3-dimethyl-4-(phosphonooxy)butanoyl]-beta-alaninamide, PCP-C didomain | | Authors: | Izore, T, Ho, Y.T.C, Kaczmarski, J.A, Gavriilidou, A, Chow, K.H, Steer, D, Goode, R.J.A, Schittenhelm, R.B, Tailhades, J, Tosin, M, Challis, G.L, Krenske, E.H, Ziemert, N, Jackson, C.J, Cryle, M.J. | | Deposit date: | 2020-11-29 | | Release date: | 2021-03-24 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structures of a non-ribosomal peptide synthetase condensation domain suggest the basis of substrate selectivity.
Nat Commun, 12, 2021
|
|
8F7H
 
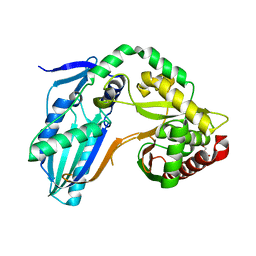 | | The condensation domain of surfactin A synthetase C variant 18b in space group P212121 | | Descriptor: | GLYCEROL, Surfactin synthetase | | Authors: | Frota, N.F, Pistofidis, A, Folger, I.B, Hilvert, D, Schmeing, M. | | Deposit date: | 2022-11-18 | | Release date: | 2023-11-22 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | High-throughput reprogramming of an NRPS condensation domain.
Nat.Chem.Biol., 2024
|
|
7JTJ
 
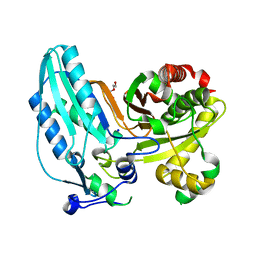 | |
4ZNM
 
 | | Crystal structure of SgcC5 protein from Streptomyces globisporus (apo form) | | Descriptor: | C-domain type II peptide synthetase, CHLORIDE ION, SODIUM ION | | Authors: | Michalska, K, Bigelow, L, Jedrzejczak, R, Babnigg, G, Lohman, J, Ma, M, Rudolf, J, Chang, C.-Y, Shen, B, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2015-05-04 | | Release date: | 2015-05-27 | | Last modified: | 2019-12-25 | | Method: | X-RAY DIFFRACTION (1.998 Å) | | Cite: | Crystal structure of SgcC5 protein from Streptomyces globisporus (apo form)
To Be Published
|
|
6CGO
 
 | | Molecular basis for condensation domain-mediated chain release from the enacyloxin polyketide synthase | | Descriptor: | Condensation domain protein, PHOSPHATE ION | | Authors: | Valentic, T.R, Tsai, S.C, Challis, G.L, Lewandowski, J.R, Kosol, S, Gallo, A, Griffiths, D, Masschelein, J.L, Jenner, M, De los Santos, E. | | Deposit date: | 2018-02-20 | | Release date: | 2019-02-27 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis for chain release from the enacyloxin polyketide synthase.
Nat.Chem., 11, 2019
|
|
7KW2
 
 | | Non-ribosomal didomain (holo-PCP-C) acceptor bound state, R2577G | | Descriptor: | 4'-PHOSPHOPANTETHEINE, PCP-C didomain | | Authors: | Izore, T, Ho, Y.T.C, Kaczmarski, J.A, Gavriilidou, A, Chow, K.H, Steer, D, Goode, R.J.A, Schittenhelm, R.B, Tailhades, J, Tosin, M, Challis, G.L, Krenske, E.H, Ziemert, N, Jackson, C.J, Cryle, M.J. | | Deposit date: | 2020-11-29 | | Release date: | 2021-03-24 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structures of a non-ribosomal peptide synthetase condensation domain suggest the basis of substrate selectivity.
Nat Commun, 12, 2021
|
|
5T7Z
 
 | | Monoclinic crystal form of the EpoB NRPS cyclization-docking bidomain from Sorangium cellulosum | | Descriptor: | EpoB | | Authors: | Dowling, D.P, Kung, Y, Croft, A.K, Taghizadeh, K, Kelly, W.L, Walsh, C.T, Drennan, C.L. | | Deposit date: | 2016-09-06 | | Release date: | 2016-11-09 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Structural elements of an NRPS cyclization domain and its intermodule docking domain.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
7X0E
 
 | | Structure of Pseudomonas NRPS protein, AmbB-TC in apo form | | Descriptor: | AMB antimetabolite synthase AmbB, N-methyl-N-[(2S,3R,4R,5R)-2,3,4,5,6-pentakis(oxidanyl)hexyl]nonanamide | | Authors: | ChuYuanKee, M, Bharath, S.R, Song, H. | | Deposit date: | 2022-02-22 | | Release date: | 2022-07-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural insights into the substrate-bound condensation domains of non-ribosomal peptide synthetase AmbB.
Sci Rep, 12, 2022
|
|
6OYF
 
 | | The structure of condensation and adenylation domains of teixobactin-producing nonribosomal peptide synthetase Txo1 serine module | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, FORMIC ACID, ... | | Authors: | Tan, K, Zhou, M, Jedrzejczak, R, Babnigg, G, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2019-05-14 | | Release date: | 2019-05-29 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structures of teixobactin-producing nonribosomal peptide synthetase condensation and adenylation domains.
Curr Res Struct Biol, 2, 2020
|
|
8G3J
 
 | |
6P4U
 
 | | The structure of condensation and adenylation domains of teixobactin-producing nonribosomal peptide synthetase Txo1 serine module in complex with Mg and AMP | | Descriptor: | ACETATE ION, ADENOSINE MONOPHOSPHATE, CHLORIDE ION, ... | | Authors: | Tan, K, Zhou, M, Jedrzejczak, R, Babnigg, G, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2019-05-28 | | Release date: | 2019-06-12 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structures of teixobactin-producing nonribosomal peptide synthetase condensation and adenylation domains.
Curr Res Struct Biol, 2, 2020
|
|
7R9X
 
 | |
