1AUA
 
 | | PHOSPHATIDYLINOSITOL TRANSFER PROTEIN SEC14P FROM SACCHAROMYCES CEREVISIAE | | Descriptor: | PHOSPHATIDYLINOSITOL TRANSFER PROTEIN SEC14P, octyl beta-D-glucopyranoside | | Authors: | Sha, B, Phillips, S.E, Bankaitis, V.A, Luo, M. | | Deposit date: | 1997-08-20 | | Release date: | 1997-12-24 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of the Saccharomyces cerevisiae phosphatidylinositol-transfer protein.
Nature, 391, 1998
|
|
1O6U
 
 | |
1R5L
 
 | | Crystal Structure of Human Alpha-Tocopherol Transfer Protein Bound to its Ligand | | Descriptor: | (2R)-2,5,7,8-TETRAMETHYL-2-[(4R,8R)-4,8,12-TRIMETHYLTRIDECYL]CHROMAN-6-OL, PROTEIN (Alpha-tocopherol transfer protein) | | Authors: | Min, K.C, Kovall, R.A, Hendrickson, W.A. | | Deposit date: | 2003-10-10 | | Release date: | 2003-11-25 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of human alpha-tocopherol transfer protein bound to its ligand: Implications for ataxia with vitamin E deficiency
Proc.Natl.Acad.Sci.USA, 100, 2003
|
|
1OIP
 
 | | The Molecular Basis of Vitamin E Retention: Structure of Human Alpha-Tocopherol Transfer Protein | | Descriptor: | (2R)-2,5,7,8-TETRAMETHYL-2-[(4R,8R)-4,8,12-TRIMETHYLTRIDECYL]CHROMAN-6-OL, ALPHA-TOCOPHEROL TRANSFER PROTEIN, SULFATE ION | | Authors: | Meier, R, Tomizaki, T, Schulze-Briese, C, Baumann, U, Stocker, A. | | Deposit date: | 2003-06-24 | | Release date: | 2004-01-14 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | The Molecular Basis of Vitamin E Retention: Structure of Human Alpha-Tocopherol Transfer Protein
J.Mol.Biol., 331, 2003
|
|
1OIZ
 
 | | The Molecular Basis of Vitamin E Retention: Structure of Human Alpha-Tocopherol Transfer Protein | | Descriptor: | ALPHA-TOCOPHEROL TRANSFER PROTEIN, FRAGMENT OF TRITON X-100 | | Authors: | Meier, R, Tomizaki, T, Schulze-Briese, C, Baumann, U, Stocker, A. | | Deposit date: | 2003-06-27 | | Release date: | 2004-01-14 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | The Molecular Basis of Vitamin E Retention: Structure of Human Alpha-Tocopherol Transfer Protein
J.Mol.Biol., 331, 2003
|
|
1OLM
 
 | |
3B7Q
 
 | | Crystal Structure of Yeast Sec14 Homolog Sfh1 in Complex with Phosphatidylcholine | | Descriptor: | (4S,7R)-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5,8-TRIOXA-4-PHOSPHAHEXACOSAN-1-AMINIUM 4-OXIDE, GLYCEROL, PHOSPHATE ION, ... | | Authors: | Ortlund, E.A, Schaaf, G, Redinbo, M.R, Bankaitis, V. | | Deposit date: | 2007-10-31 | | Release date: | 2008-02-19 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Functional anatomy of phospholipid binding and regulation of phosphoinositide homeostasis by proteins of the sec14 superfamily
Mol.Cell, 29, 2008
|
|
3B74
 
 | | Crystal Structure of Yeast Sec14 Homolog Sfh1 in Complex with Phosphatidylethanolamine | | Descriptor: | 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, Uncharacterized protein YKL091C | | Authors: | Ortlund, E.A, Schaaf, G, Redinbo, M.R, Bankaitis, V. | | Deposit date: | 2007-10-30 | | Release date: | 2008-02-19 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Functional anatomy of phospholipid binding and regulation of phosphoinositide homeostasis by proteins of the sec14 superfamily
Mol.Cell, 29, 2008
|
|
3B7N
 
 | | Crystal Structure of Yeast Sec14 Homolog Sfh1 in Complex with Phosphatidylinositol | | Descriptor: | (1R)-2-{[(S)-hydroxy{[(1S,2R,3R,4S,5S,6R)-2,3,4,5,6-pentahydroxycyclohexyl]oxy}phosphoryl]oxy}-1-[(octadecanoyloxy)methyl]ethyl (9Z)-octadec-9-enoate, ACETATE ION, PHOSPHATE ION, ... | | Authors: | Ortlund, E.A, Schaaf, G, Redinbo, M.R, Bankaitis, V. | | Deposit date: | 2007-10-31 | | Release date: | 2008-02-19 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Functional anatomy of phospholipid binding and regulation of phosphoinositide homeostasis by proteins of the sec14 superfamily
Mol.Cell, 29, 2008
|
|
3B7Z
 
 | | Crystal Structure of Yeast Sec14 Homolog Sfh1 in Complex with Phosphatidylcholine or Phosphatidylinositol | | Descriptor: | (1R)-2-{[(S)-hydroxy{[(1S,2R,3R,4S,5S,6R)-2,3,4,5,6-pentahydroxycyclohexyl]oxy}phosphoryl]oxy}-1-[(octadecanoyloxy)methyl]ethyl (9Z)-octadec-9-enoate, (4S,7R)-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5,8-TRIOXA-4-PHOSPHAHEXACOSAN-1-AMINIUM 4-OXIDE, Uncharacterized protein YKL091C | | Authors: | Ortlund, E.A, Schaaf, G, Redinbo, M.R, Bankaitis, V. | | Deposit date: | 2007-10-31 | | Release date: | 2008-02-19 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Functional anatomy of phospholipid binding and regulation of phosphoinositide homeostasis by proteins of the sec14 superfamily
Mol.Cell, 29, 2008
|
|
3HY5
 
 | | Crystal structure of CRALBP | | Descriptor: | L(+)-TARTARIC ACID, RETINAL, Retinaldehyde-binding protein 1 | | Authors: | Stocker, A, He, X, Lobsiger, J. | | Deposit date: | 2009-06-22 | | Release date: | 2009-10-20 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.04 Å) | | Cite: | Bothnia dystrophy is caused by domino-like rearrangements in cellular retinaldehyde-binding protein mutant R234W.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
3HX3
 
 | | Crystal structure of CRALBP mutant R234W | | Descriptor: | RETINAL, Retinaldehyde-binding protein 1 | | Authors: | Stocker, A, He, X, Lobsiger, J. | | Deposit date: | 2009-06-19 | | Release date: | 2009-10-20 | | Last modified: | 2021-10-13 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Bothnia dystrophy is caused by domino-like rearrangements in cellular retinaldehyde-binding protein mutant R234W.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
3Q8G
 
 | | Resurrection of a functional phosphatidylinositol transfer protein from a pseudo-Sec14 scaffold by directed evolution | | Descriptor: | 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, CRAL-TRIO domain-containing protein YKL091C, GLYCEROL | | Authors: | Ortlund, E.A, Schaaf, G, Bankaitis, V.A. | | Deposit date: | 2011-01-06 | | Release date: | 2011-02-23 | | Last modified: | 2023-05-31 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Resurrection of a functional phosphatidylinositol transfer protein from a pseudo-Sec14 scaffold by directed evolution.
Mol.Biol.Cell, 22, 2011
|
|
4FMM
 
 | | Dimeric Sec14 family homolog 3 from Saccharomyces cerevisiae presents some novel features of structure that lead to a surprising "dimer-monomer" state change induced by substrate binding | | Descriptor: | GLYCEROL, MAGNESIUM ION, Phosphatidylinositol transfer protein PDR16 | | Authors: | Yuan, Y, Zhao, W, Wang, X, Gao, Y, Niu, L, Teng, M. | | Deposit date: | 2012-06-18 | | Release date: | 2013-02-27 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Dimeric Sfh3 has structural changes in its binding pocket that are associated with a dimer-monomer state transformation induced by substrate binding.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
3W67
 
 | | Crystal structure of mouse alpha-tocopherol transfer protein in complex with alpha-tocopherol and phosphatidylinositol-(3,4)-bisphosphate | | Descriptor: | (2R)-2,5,7,8-TETRAMETHYL-2-[(4R,8R)-4,8,12-TRIMETHYLTRIDECYL]CHROMAN-6-OL, (2R)-3-{[(S)-hydroxy{[(1S,2R,3R,4S,5S,6S)-2,3,6-trihydroxy-4,5-bis(phosphonooxy)cyclohexyl]oxy}phosphoryl]oxy}propane-1,2-diyl dibutanoate, Alpha-tocopherol transfer protein | | Authors: | Ohto, U, Satow, Y. | | Deposit date: | 2013-02-11 | | Release date: | 2013-05-01 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Impaired alpha-TTP-PIPs interaction underlies familial vitamin E deficiency
Science, 340, 2013
|
|
3W68
 
 | | Crystal structure of mouse alpha-tocopherol transfer protein in complex with alpha-tocopherol and phosphatidylinositol-(4,5)-bisphosphate | | Descriptor: | (2R)-2,5,7,8-TETRAMETHYL-2-[(4R,8R)-4,8,12-TRIMETHYLTRIDECYL]CHROMAN-6-OL, (2R)-3-{[(R)-HYDROXY{[(1R,2R,3S,4R,5R,6S)-2,3,6-TRIHYDROXY-4,5-BIS(PHOSPHONOOXY)CYCLOHEXYL]OXY}PHOSPHORYL]OXY}PROPANE-1 ,2-DIYL DIBUTANOATE, (2R)-3-{[(S)-{[(2S,3R,5S,6S)-2,6-DIHYDROXY-3,4,5-TRIS(PHOSPHONOOXY)CYCLOHEXYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-2-(1-HYDROXY BUTOXY)PROPYL BUTYRATE, ... | | Authors: | Ohto, U, Satow, Y. | | Deposit date: | 2013-02-11 | | Release date: | 2013-05-01 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Impaired alpha-TTP-PIPs interaction underlies familial vitamin E deficiency
Science, 340, 2013
|
|
4J7Q
 
 | | Crystal structure of Saccharomyces cerevisiae Sfh3 complexed with phosphatidylinositol | | Descriptor: | (1R)-2-{[(S)-hydroxy{[(1S,2R,3R,4S,5S,6R)-2,3,4,5,6-pentahydroxycyclohexyl]oxy}phosphoryl]oxy}-1-[(octadecanoyloxy)methyl]ethyl (9Z)-octadec-9-enoate, Phosphatidylinositol transfer protein PDR16 | | Authors: | Yang, H, Im, Y.J. | | Deposit date: | 2013-02-14 | | Release date: | 2013-07-03 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structural determinants for phosphatidylinositol recognition by Sfh3 and substrate-induced dimer-monomer transition during lipid transfer cycles.
Febs Lett., 587, 2013
|
|
4J7P
 
 | | Crystal structure of Saccharomyces cerevisiae Sfh3 | | Descriptor: | Phosphatidylinositol transfer protein PDR16 | | Authors: | Yang, H, Im, Y.J. | | Deposit date: | 2013-02-14 | | Release date: | 2013-07-03 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural determinants for phosphatidylinositol recognition by Sfh3 and substrate-induced dimer-monomer transition during lipid transfer cycles.
Febs Lett., 587, 2013
|
|
4CIZ
 
 | | Crystal structure of the complex of the Cellular Retinal Binding Protein with 9-cis-retinal | | Descriptor: | L(+)-TARTARIC ACID, RETINAL, RETINALDEHYDE-BINDING PROTEIN 1 | | Authors: | Bolze, C.S, Helbling, R.E, Owen, R.L, Pearson, A.R, Pompidor, G, Dworkowski, F, Fuchs, M.R, Furrer, J, Golczak, M, Palczewski, K, Cascella, M, Stocker, A. | | Deposit date: | 2013-12-18 | | Release date: | 2014-01-08 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.403 Å) | | Cite: | Human Cellular Retinaldehyde-Binding Protein Has Secondary Thermal 9-Cis-Retinal Isomerase Activity.
J.Am.Chem.Soc., 136, 2014
|
|
4CJ6
 
 | | Crystal structure of the complex of the Cellular Retinal Binding Protein Mutant R234W with 9-cis-retinal | | Descriptor: | RETINAL, RETINALDEHYDE-BINDING PROTEIN 1 | | Authors: | Bolze, C.S, Helbling, R.E, Owen, R.L, Pearson, A.R, Pompidor, G, Dworkowski, F, Fuchs, M.R, Furrer, J, Golczak, M, Palczewski, K, Cascella, M, Stocker, A. | | Deposit date: | 2013-12-19 | | Release date: | 2014-01-08 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.896 Å) | | Cite: | Human Cellular Retinaldehyde-Binding Protein Has Secondary Thermal 9-Cis-Retinal Isomerase Activity.
J.Am.Chem.Soc., 136, 2014
|
|
4TLG
 
 | | Crystal structure of SEC14-like protein 4 (SEC14L4) | | Descriptor: | 1,2-ETHANEDIOL, SEC14-like protein 4, UNDECANOIC ACID | | Authors: | Vollmar, M, Kopec, J, Kiyani, W, Shrestha, L, Sorrell, F, Krojer, T, Williams, E, Burgess-Brown, N, von Delft, F, Arrowsmith, C, Edwards, A, Bountra, C, Yue, W.W. | | Deposit date: | 2014-05-29 | | Release date: | 2014-07-02 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Crystal structure of SEC14-like protein 4 (SEC14L4)
To Be Published
|
|
4M8Z
 
 | |
4UYB
 
 | | Crystal structure of SEC14-like protein 3 | | Descriptor: | 1,2-ETHANEDIOL, SEC14-LIKE PROTEIN 3, UNKNOWN LIGAND | | Authors: | Kopec, J, Goubin, S, Krojer, T, Burgess-Brown, N, von Delft, F, Arrowsmith, C, Edwards, A, Bountra, C, Yue, W.W. | | Deposit date: | 2014-08-29 | | Release date: | 2014-09-24 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal Structure of Sec14-Like Protein 3
To be Published
|
|
4OMJ
 
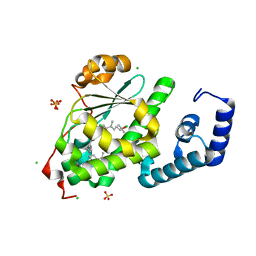 | | Crystal structure of SPF bound to 2,3-oxidosqualene | | Descriptor: | (3S)-2,2-dimethyl-3-[(3E,7E,11E,15E)-3,7,12,16,20-pentamethylhenicosa-3,7,11,15,19-pentaen-1-yl]oxirane, CHLORIDE ION, SEC14-like protein 2, ... | | Authors: | Christen, M, Marcaida, M.J, Lamprakis, C, Cascella, M, Stocker, A. | | Deposit date: | 2014-01-27 | | Release date: | 2015-04-15 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural insights on cholesterol endosynthesis: Binding of squalene and 2,3-oxidosqualene to supernatant protein factor.
J.Struct.Biol., 190, 2015
|
|
4OMK
 
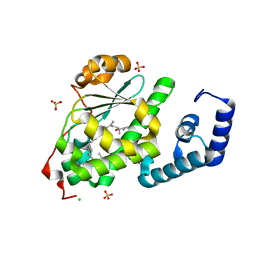 | | Crystal structure of SPF bound to squalene | | Descriptor: | (6E,10E,14E,18E)-2,6,10,15,19,23-hexamethyltetracosa-2,6,10,14,18,22-hexaene, CHLORIDE ION, ISOPROPYL ALCOHOL, ... | | Authors: | Christen, M, Marcaida, M.J, Lamprakis, C, Cascella, M, Stocker, A. | | Deposit date: | 2014-01-27 | | Release date: | 2015-04-15 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural insights on cholesterol endosynthesis: Binding of squalene and 2,3-oxidosqualene to supernatant protein factor.
J.Struct.Biol., 190, 2015
|
|
