1ORQ
 
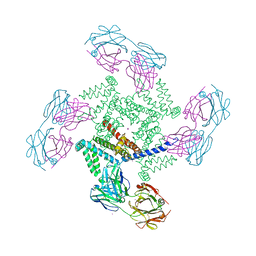 | | X-ray structure of a voltage-dependent potassium channel in complex with an Fab | | Descriptor: | 6E1 Fab heavy chain, 6E1 Fab light chain, CADMIUM ION, ... | | Authors: | Jiang, Y, Lee, A, Chen, J, Ruta, V, Cadene, M, Chait, B.T, MacKinnon, R. | | Deposit date: | 2003-03-14 | | Release date: | 2003-05-06 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | X-ray structure of a voltage-dependent K+ channel
Nature, 423, 2003
|
|
1ORS
 
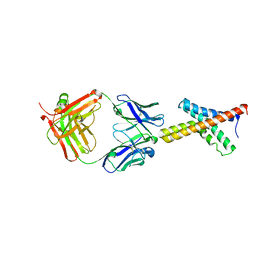 | | X-ray structure of the KvAP potassium channel voltage sensor in complex with an Fab | | Descriptor: | 33H1 Fab heavy chain, 33H1 Fab light chain, potassium channel | | Authors: | Jiang, Y, Lee, A, Chen, J, Ruta, V, Cadene, M, Chait, B.T, MacKinnon, R. | | Deposit date: | 2003-03-14 | | Release date: | 2003-05-06 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | X-ray structure of a voltage-dependent K+ channel
Nature, 423, 2003
|
|
2A0L
 
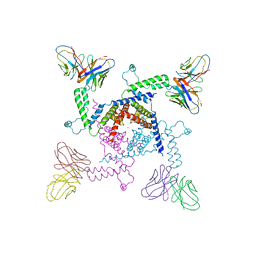 | | Crystal structure of KvAP-33H1 Fv complex | | Descriptor: | 33H1 Fv fragment, POTASSIUM ION, Voltage-gated potassium channel | | Authors: | Lee, S.Y, Lee, A, Chen, J, Mackinnon, R. | | Deposit date: | 2005-06-16 | | Release date: | 2005-11-01 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (3.9 Å) | | Cite: | Structure of the KvAP voltage-dependent K+ channel and its dependence on the lipid membrane.
Proc.Natl.Acad.Sci.Usa, 102, 2005
|
|
2A79
 
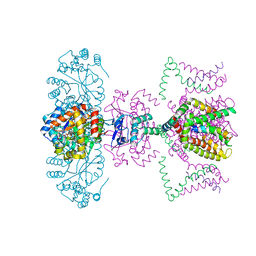 | | Mammalian Shaker Kv1.2 potassium channel- beta subunit complex | | Descriptor: | NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, POTASSIUM ION, Potassium voltage-gated channel subfamily A member 2, ... | | Authors: | Long, S.B, Campbell, E.B, MacKinnon, R. | | Deposit date: | 2005-07-05 | | Release date: | 2005-07-12 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structure of a mammalian voltage-dependent Shaker family K+ channel.
Science, 309, 2005
|
|
2KYH
 
 | |
2ZD9
 
 | |
3BEH
 
 | |
3J5P
 
 | |
3J5Q
 
 | | Structure of TRPV1 ion channel in complex with DkTx and RTX determined by single particle electron cryo-microscopy | | Descriptor: | Kappa-theraphotoxin-Cg1a 1, Transient receptor potential cation channel subfamily V member 1 | | Authors: | Liao, M, Cao, E, Julius, D, Cheng, Y. | | Deposit date: | 2013-10-28 | | Release date: | 2013-12-04 | | Last modified: | 2022-11-02 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | TRPV1 structures in distinct conformations reveal activation mechanisms.
Nature, 504, 2013
|
|
3J5R
 
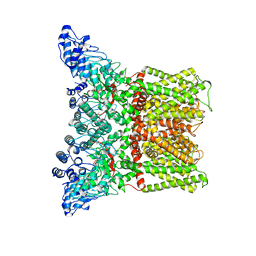 | |
3J8H
 
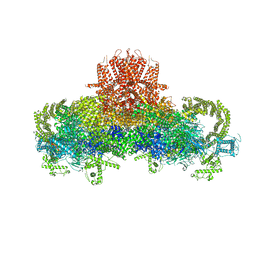 | | Structure of the rabbit ryanodine receptor RyR1 in complex with FKBP12 at 3.8 Angstrom resolution | | Descriptor: | Peptidyl-prolyl cis-trans isomerase FKBP1A, Ryanodine receptor 1, ZINC ION | | Authors: | Yan, Z, Bai, X, Yan, C, Wu, J, Scheres, S.H.W, Shi, Y, Yan, N. | | Deposit date: | 2014-10-26 | | Release date: | 2014-12-10 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structure of the rabbit ryanodine receptor RyR1 at near-atomic resolution.
Nature, 517, 2015
|
|
3J9J
 
 | | Structure of the capsaicin receptor, TRPV1, determined by single particle electron cryo-microscopy | | Descriptor: | Transient receptor potential cation channel subfamily V member 1 | | Authors: | Wang, R.Y.-R, Barad, B.A, Fraser, J.S, DiMaio, F. | | Deposit date: | 2015-02-02 | | Release date: | 2015-09-02 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (3.275 Å) | | Cite: | EMRinger: side chain-directed model and map validation for 3D cryo-electron microscopy.
Nat.Methods, 12, 2015
|
|
3J9P
 
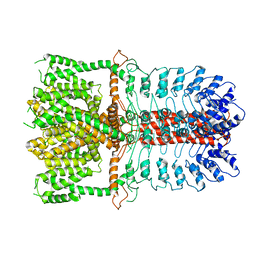 | | Structure of the TRPA1 ion channel determined by electron cryo-microscopy | | Descriptor: | Maltose-binding periplasmic protein, Transient receptor potential cation channel subfamily A member 1 chimera | | Authors: | Paulsen, C.E, Armache, J.-P, Gao, Y, Cheng, Y, Julius, D. | | Deposit date: | 2015-02-14 | | Release date: | 2015-04-08 | | Last modified: | 2023-02-01 | | Method: | ELECTRON MICROSCOPY (4.24 Å) | | Cite: | Structure of the TRPA1 ion channel suggests regulatory mechanisms.
Nature, 520, 2015
|
|
3JAV
 
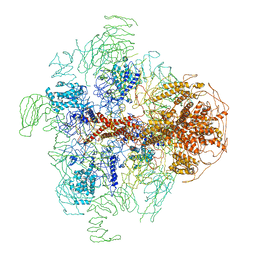 | | Structure of full-length IP3R1 channel in the apo-state determined by single particle cryo-EM | | Descriptor: | Inositol 1,4,5-trisphosphate receptor type 1 | | Authors: | Fan, G, Baker, M.L, Wang, Z, Baker, M.R, Sinyagovskiy, P.A, Chiu, W, Ludtke, S.J, Serysheva, I.I. | | Deposit date: | 2015-06-30 | | Release date: | 2015-10-07 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (4.7 Å) | | Cite: | Gating machinery of InsP3R channels revealed by electron cryomicroscopy.
Nature, 527, 2015
|
|
3JBR
 
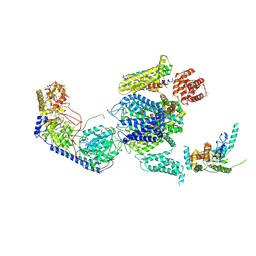 | | Cryo-EM structure of the rabbit voltage-gated calcium channel Cav1.1 complex at 4.2 angstrom | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Voltage-dependent L-type calcium channel subunit alpha-1S, ... | | Authors: | Wu, J.P, Yan, Z, Yan, N. | | Deposit date: | 2015-09-29 | | Release date: | 2015-12-30 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Structure of the voltage-gated calcium channel Cav1.1 complex
Science, 350, 2015
|
|
3LNM
 
 | | F233W mutant of the Kv2.1 paddle-Kv1.2 chimera channel | | Descriptor: | (1R)-2-{[(S)-{[(2S)-2,3-dihydroxypropyl]oxy}(hydroxy)phosphoryl]oxy}-1-[(hexadecanoyloxy)methyl]ethyl (9Z)-octadec-9-enoate, F233W mutant of the Kv2.1 paddle-Kv1.2 chimera, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Tao, X, Lee, A, Limapichat, W, Dougherty, D.A, MacKinnon, R. | | Deposit date: | 2010-02-02 | | Release date: | 2010-04-14 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | A gating charge transfer center in voltage sensors.
Science, 328, 2010
|
|
3LUT
 
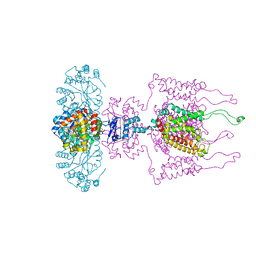 | | A Structural Model for the Full-length Shaker Potassium Channel Kv1.2 | | Descriptor: | NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, POTASSIUM ION, Potassium voltage-gated channel subfamily A member 2, ... | | Authors: | Chen, X, Ni, F, Wang, Q, Ma, J. | | Deposit date: | 2010-02-18 | | Release date: | 2010-06-23 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structure of the full-length Shaker potassium channel Kv1.2 by normal-mode-based X-ray crystallographic refinement.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
3RVY
 
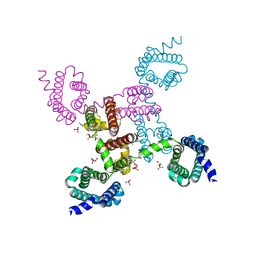 | | Crystal structure of the NavAb voltage-gated sodium channel (Ile217Cys, 2.7 A) | | Descriptor: | 1,2-DIMYRISTOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, Ion transport protein | | Authors: | Payandeh, J, Scheuer, T, Zheng, N, Catterall, W.A. | | Deposit date: | 2011-05-06 | | Release date: | 2011-07-13 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The crystal structure of a voltage-gated sodium channel.
Nature, 475, 2011
|
|
3RVZ
 
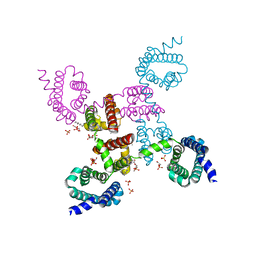 | | Crystal structure of the NavAb voltage-gated sodium channel (Ile217Cys, 2.8 A) | | Descriptor: | 1,2-DIMYRISTOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, Ion transport protein | | Authors: | Payandeh, J, Scheuer, T, Zheng, N, Catterall, W.A. | | Deposit date: | 2011-05-06 | | Release date: | 2011-07-13 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The crystal structure of a voltage-gated sodium channel.
Nature, 475, 2011
|
|
3RW0
 
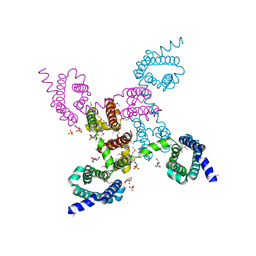 | | Crystal structure of the NavAb voltage-gated sodium channel (Met221Cys, 2.95 A) | | Descriptor: | 1,2-DIMYRISTOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, Ion transport protein | | Authors: | Payandeh, J, Scheuer, T, Zheng, N, Catterall, W.A. | | Deposit date: | 2011-05-06 | | Release date: | 2011-07-13 | | Last modified: | 2011-07-27 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | The crystal structure of a voltage-gated sodium channel.
Nature, 475, 2011
|
|
3WKV
 
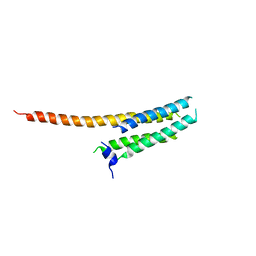 | | Voltage-gated proton channel: VSOP/Hv1 chimeric channel | | Descriptor: | Ion channel | | Authors: | Takeshita, K, Sakata, S, Yamashita, E, Fujiwara, Y, Kawanabe, A, Kurokawa, T, Okochi, Y, Matsuda, M, Narita, H, Okamura, Y, Nakagawa, A. | | Deposit date: | 2013-10-31 | | Release date: | 2014-03-05 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3.453 Å) | | Cite: | X-ray crystal structure of voltage-gated proton channel.
Nat.Struct.Mol.Biol., 21, 2014
|
|
3ZJZ
 
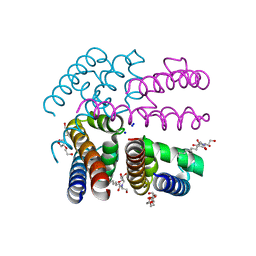 | | Open-form NavMS Sodium Channel Pore (with C-terminal Domain) | | Descriptor: | DODECAETHYLENE GLYCOL, HEGA-10, ION TRANSPORT PROTEIN, ... | | Authors: | Bagneris, C, Naylor, C.E, Wallace, B.A. | | Deposit date: | 2013-01-21 | | Release date: | 2013-10-02 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.92 Å) | | Cite: | Role of the C-Terminal Domain in the Structure and Function of Tetrameric Sodium Channels.
Nat.Commun., 4, 2013
|
|
4BGN
 
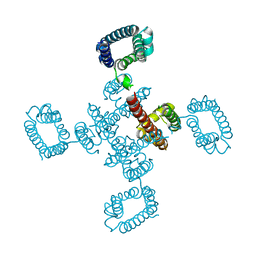 | | cryo-EM structure of the NavCt voltage-gated sodium channel | | Descriptor: | VOLTAGE-GATED SODIUM CHANNEL | | Authors: | Tsai, C.J, Tani, K, Irie, K, Hiroaki, Y, Shimomura, T, Mcmillan, D.G, Cook, G.M, Schertler, G, Fujiyoshi, Y, Li, X.D. | | Deposit date: | 2013-03-28 | | Release date: | 2013-07-10 | | Last modified: | 2023-12-20 | | Method: | ELECTRON CRYSTALLOGRAPHY (9 Å) | | Cite: | Two Alternative Conformations of a Voltage-Gated Sodium Channel.
J.Mol.Biol., 425, 2013
|
|
4CBC
 
 | |
4CHV
 
 | | The electron crystallography structure of the cAMP-bound potassium channel MloK1 | | Descriptor: | CYCLIC NUCLEOTIDE-GATED POTASSIUM CHANNEL MLL3241, POTASSIUM ION | | Authors: | Kowal, J, Chami, M, Baumgartner, P, Arheit, M, Chiu, P.L, Rangl, M, Scheuring, S, Schroeder, G.F, Nimigean, C.M, Stahlberg, H. | | Deposit date: | 2013-12-04 | | Release date: | 2014-01-15 | | Last modified: | 2019-04-24 | | Method: | ELECTRON CRYSTALLOGRAPHY (7 Å) | | Cite: | Ligand-Induced Structural Changes in the Cyclic Nucleotide-Modulated Potassium Channel Mlok1
Nat.Commun., 5, 2014
|
|
