8X39
 
 | |
8X3A
 
 | |
2MTE
 
 | | Solution structure of Doc48S | | Descriptor: | CALCIUM ION, Cellulose 1,4-beta-cellobiosidase (reducing end) CelS | | Authors: | Chen, C, Feng, Y. | | Deposit date: | 2014-08-18 | | Release date: | 2014-10-15 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Revisiting the NMR solution structure of the Cel48S type-I dockerin module from Clostridium thermocellum reveals a cohesin-primed conformation.
J.Struct.Biol., 188, 2014
|
|
1DAV
 
 | | SOLUTION STRUCTURE OF THE TYPE I DOCKERIN DOMAIN FROM THE CLOSTRIDIUM THERMOCELLUM CELLULOSOME (20 STRUCTURES) | | Descriptor: | CALCIUM ION, ENDOGLUCANASE SS | | Authors: | Lytle, B.L, Volkman, B.F, Westler, W.M, Heckman, M.P, Wu, J.H.D. | | Deposit date: | 1999-10-31 | | Release date: | 2001-04-04 | | Last modified: | 2021-11-03 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a type I dockerin domain, a novel prokaryotic, extracellular calcium-binding domain.
J.Mol.Biol., 307, 2001
|
|
1DAQ
 
 | | SOLUTION STRUCTURE OF THE TYPE I DOCKERIN DOMAIN FROM THE CLOSTRIDIUM THERMOCELLUM CELLULOSOME (MINIMIZED AVERAGE STRUCTURE) | | Descriptor: | CALCIUM ION, ENDOGLUCANASE SS | | Authors: | Lytle, B.L, Volkman, B.F, Westler, W.M, Heckman, M.P, Wu, J.H.D. | | Deposit date: | 1999-10-31 | | Release date: | 2001-04-04 | | Last modified: | 2021-11-03 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a type I dockerin domain, a novel prokaryotic, extracellular calcium-binding domain.
J.Mol.Biol., 307, 2001
|
|
6KG9
 
 | | Solution structure of CaDoc0917 from Clostridium acetobutylicum | | Descriptor: | And cellulose-binding endoglucanase family 9 CelL ortholog dockerin domain, CALCIUM ION | | Authors: | Feng, Y, Yao, X. | | Deposit date: | 2019-07-11 | | Release date: | 2020-07-08 | | Last modified: | 2020-11-04 | | Method: | SOLUTION NMR | | Cite: | Discovery and mechanism of a pH-dependent dual-binding-site switch in the interaction of a pair of protein modules.
Sci Adv, 6, 2020
|
|
5G5D
 
 | | Crystal Structure of the CohScaC2-XDocCipA type II complex from Clostridium thermocellum | | Descriptor: | CALCIUM ION, CELLULOSOMAL-SCAFFOLDING PROTEIN A, CELLULOSOME ANCHORING PROTEIN COHESIN REGION | | Authors: | Carvalho, A.L, A Bras, J.L, Najmudin, S.H, Pinheiro, B.A, Fontes, C.M.G.A. | | Deposit date: | 2016-05-23 | | Release date: | 2017-04-05 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Diverse specificity of cellulosome attachment to the bacterial cell surface.
Sci Rep, 6, 2016
|
|
4FL4
 
 | | Scaffoldin conformation and dynamics revealed by a ternary complex from the Clostridium thermocellum cellulosome | | Descriptor: | CALCIUM ION, Cellulosome anchoring protein cohesin region, Glycoside hydrolase family 9, ... | | Authors: | Currie, M.A, Adams, J.J, Faucher, F, Bayer, E.A, Jia, Z, Smith, S.P, Montreal-Kingston Bacterial Structural Genomics Initiative (BSGI) | | Deposit date: | 2012-06-14 | | Release date: | 2012-06-27 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Scaffoldin Conformation and Dynamics Revealed by a Ternary Complex from the Clostridium thermocellum Cellulosome.
J.Biol.Chem., 287, 2012
|
|
1OHZ
 
 | | Cohesin-Dockerin complex from the cellulosome of Clostridium thermocellum | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, CELLULOSOMAL SCAFFOLDING PROTEIN A, ... | | Authors: | Carvalho, A.L, Dias, F.M.V, Prates, J.A.M, Ferreira, L.M.A, Gilbert, H.J, Davies, G.J, Romao, M.J, Fontes, C.M.G.A. | | Deposit date: | 2003-06-04 | | Release date: | 2003-11-20 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Cellulosome Assembly Revealed by the Crystal Structure of the Cohesin-Dockerin Complex
Proc.Natl.Acad.Sci.USA, 100, 2003
|
|
3UL4
 
 | | Crystal structure of Coh-OlpA(Cthe_3080)-Doc918(Cthe_0918) complex: A novel type I Cohesin-Dockerin complex from Clostridium thermocellum ATTC 27405 | | Descriptor: | CALCIUM ION, Cellulosome enzyme, dockerin type I, ... | | Authors: | Alves, V.D, Carvalho, A.L, Najmudin, S.H, Bras, J, Prates, J.A.M, Fontes, C.M.G.A. | | Deposit date: | 2011-11-10 | | Release date: | 2012-11-28 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Novel Clostridium thermocellum Type I Cohesin-Dockerin Complexes Reveal a Single Binding Mode.
J.Biol.Chem., 287, 2012
|
|
2JNK
 
 | |
3KCP
 
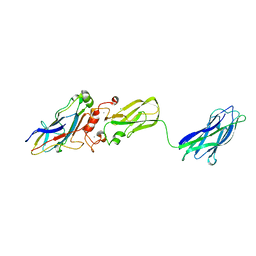 | | Crystal structure of interacting Clostridium thermocellum multimodular components | | Descriptor: | CALCIUM ION, CHLORIDE ION, Cellulosomal-scaffolding protein A, ... | | Authors: | Adams, J.J, Currie, M.A, Bayer, E.A, Jia, Z, Smith, S.P. | | Deposit date: | 2009-10-21 | | Release date: | 2010-02-09 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Insights into Higher-Order Organization of the Cellulosome Revealed by a Dissect-and-Build Approach: Crystal Structure of Interacting Clostridium thermocellum Multimodular Components
J.Mol.Biol., 396, 2010
|
|
4UYQ
 
 | |
4UYP
 
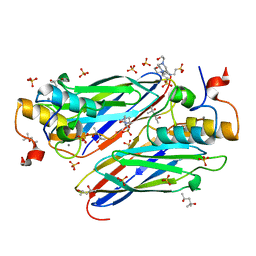 | | High resolution structure of the third cohesin ScaC in complex with the ScaB dockerin with a mutation in the N-terminal helix (IN to SI) from Acetivibrio cellulolyticus displaying a type I interaction. | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CALCIUM ION, ... | | Authors: | Cameron, K, Alves, V.D, Bule, P, Ferreira, L.M.A, Fontes, C.M.G.A, Najmudin, S. | | Deposit date: | 2014-09-02 | | Release date: | 2015-04-15 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Cell-surface Attachment of Bacterial Multienzyme Complexes Involves Highly Dynamic Protein-Protein Anchors.
J. Biol. Chem., 290, 2015
|
|
2B59
 
 | |
2OZN
 
 | | The Cohesin-Dockerin Complex of NagJ and NagH from Clostridium perfringens | | Descriptor: | CALCIUM ION, CHLORIDE ION, Hyalurononglucosaminidase, ... | | Authors: | Adams, J.J, Boraston, A, Smith, S.P. | | Deposit date: | 2007-02-26 | | Release date: | 2008-05-06 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural basis of Clostridium perfringens toxin complex formation.
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
2CCL
 
 | | THE S45A, T46A MUTANT OF THE TYPE I COHESIN-DOCKERIN COMPLEX FROM THE CELLULOSOME OF CLOSTRIDIUM THERMOCELLUM | | Descriptor: | CALCIUM ION, CELLULOSOMAL SCAFFOLDING PROTEIN A, ENDO-1,4-BETA-XYLANASE Y, ... | | Authors: | Carvalho, A.L, Dias, F.M.V, Prates, J.A.M, Ferreira, L.M.A, Gilbert, H.J, Davies, G.J, Romao, M.J, Fontes, C.M.G.A. | | Deposit date: | 2006-01-16 | | Release date: | 2007-02-13 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Evidence for a Dual Binding Mode of Dockerin Modules to Cohesins.
Proc.Natl.Acad.Sci.USA, 104, 2007
|
|
1CLC
 
 | |
4CJ1
 
 | | Crystal structure of CelD in complex with affitin H3 | | Descriptor: | CALCIUM ION, ENDOGLUCANASE D, GLYCEROL, ... | | Authors: | Correa, A, Pacheco, S, Mechaly, A.E, Obal, G, Behar, G, Mouratou, B, Oppezzo, P, Alzari, P.M, Pecorari, F. | | Deposit date: | 2013-12-18 | | Release date: | 2014-05-21 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Potent and Specific Inhibition of Glycosidases by Small Artificial Binding Proteins (Affitins)
Plos One, 9, 2014
|
|
4CJ0
 
 | | Crystal structure of CelD in complex with affitin E12 | | Descriptor: | CALCIUM ION, E12 AFFITIN, ENDOGLUCANASE D, ... | | Authors: | Correa, A, Pacheco, S, Mechaly, A.E, Obal, G, Behar, G, Mouratou, B, Oppezzo, P, Alzari, P.M, Pecorari, F. | | Deposit date: | 2013-12-18 | | Release date: | 2014-05-21 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Potent and Specific Inhibition of Glycosidases by Small Artificial Binding Proteins (Affitins)
Plos One, 9, 2014
|
|
4DH2
 
 | | Crystal structure of Coh-OlpC(Cthe_0452)-Doc435(Cthe_0435) complex: A novel type I Cohesin-Dockerin complex from Clostridium thermocellum ATTC 27405 | | Descriptor: | CALCIUM ION, Cellulosome anchoring protein cohesin region, Dockerin type 1, ... | | Authors: | Alves, V.D, Carvalho, A.L, Najmudin, S.H, Bras, J, Prates, J.A.M, Fontes, C.M.G.A. | | Deposit date: | 2012-01-27 | | Release date: | 2012-11-28 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Novel Clostridium thermocellum Type I Cohesin-Dockerin Complexes Reveal a Single Binding Mode.
J.Biol.Chem., 287, 2012
|
|
2VN6
 
 | | The Clostridium cellulolyticum dockerin displays a dual binding mode for its cohesin partner | | Descriptor: | CALCIUM ION, ENDOGLUCANASE A, SCAFFOLDING PROTEIN | | Authors: | Pinheiro, B.A, Prates, J.A.M, Proctor, M.R, Gilbert, H.J, Davies, G.J, Money, V.A, Martinez-Fleites, C, Bayer, E.A, Fontes, C.M.G.A, Fierobe, H.P. | | Deposit date: | 2008-01-31 | | Release date: | 2008-05-13 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | The Clostridium Cellulolyticum Dockerin Displays a Dual Binding Mode for its Cohesin Partner.
J.Biol.Chem., 283, 2008
|
|
2VN5
 
 | | The Clostridium cellulolyticum dockerin displays a dual binding mode for its cohesin partner | | Descriptor: | CALCIUM ION, ENDOGLUCANASE A, SCAFFOLDING PROTEIN | | Authors: | Pinheiro, B.A, Prates, J.A.M, Proctor, M.R, Gilbert, H.J, Davies, G.J, Money, V.A, Martinez-Fleites, C, Bayer, E.A, Fontes, C.M.G.A, Fierobe, H.P. | | Deposit date: | 2008-01-31 | | Release date: | 2008-05-20 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The Clostridium Cellulolyticum Dockerin Displays a Dual Binding Mode for its Cohesin Partner.
J.Biol.Chem., 283, 2008
|
|
2Y3N
 
 | | Type II cohesin-dockerin domain from Bacteroides cellolosolvens | | Descriptor: | CALCIUM ION, CELLULOSOMAL FAMILY-48 PROCESSIVE GLYCOSIDE HYDROLASE, CELLULOSOMAL SCAFFOLDIN | | Authors: | Viegas, A, Carvalho, A.L, Romao, M.J, Prates, J.A.M, Fontes, C.M.G.A. | | Deposit date: | 2010-12-22 | | Release date: | 2012-01-11 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Type II Cohesin-Dockerin Domain from Bacteroides Cellolosolvens
To be Published
|
|
2YIK
 
 | | Catalytic domain of Clostridium thermocellum CelT | | Descriptor: | CALCIUM ION, ENDOGLUCANASE, ZINC ION | | Authors: | Tsai, J.-Y, Kesavulu, M.M, Hsiao, C.-D. | | Deposit date: | 2011-05-16 | | Release date: | 2012-02-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of the Catalytic Domain of the Clostridium Thermocellum Cellulase Celt
Acta Crystallogr.,Sect.D, 68, 2012
|
|
