4V06
 
 | | Crystal structure of human tryptophan hydroxylase 2 (TPH2), catalytic domain | | Descriptor: | FE (III) ION, IMIDAZOLE, TRYPTOPHAN 5-HYDROXYLASE 2 | | Authors: | Kopec, J, Oberholzer, A, Fitzpatrick, F, Newman, J, Tallant, C, Kiyani, W, Shrestha, L, Burgess-Brown, N, von Delft, F, Arrowsmith, C, Edwards, A, Bountra, C, Yue, W.W. | | Deposit date: | 2014-09-11 | | Release date: | 2014-10-15 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.63 Å) | | Cite: | Crystal Structure of Human Tryptophane Hydroxylase 2 (Tph2), Catalytic Domain
To be Published
|
|
1DMW
 
 | | CRYSTAL STRUCTURE OF DOUBLE TRUNCATED HUMAN PHENYLALANINE HYDROXYLASE WITH BOUND 7,8-DIHYDRO-L-BIOPTERIN | | Descriptor: | 7,8-DIHYDROBIOPTERIN, FE (III) ION, PHENYLALANINE HYDROXYLASE | | Authors: | Erlandsen, H, Stevens, R.C, Flatmark, T. | | Deposit date: | 1999-12-15 | | Release date: | 2000-03-24 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure and site-specific mutagenesis of pterin-bound human phenylalanine hydroxylase.
Biochemistry, 39, 2000
|
|
5L01
 
 | | Tryptophan 5-hydroxylase in complex with inhibitor (3~{S})-8-[2-azanyl-6-[(1~{R})-1-(4-chloranyl-2-phenyl-phenyl)-2,2,2-tris(fluoranyl)ethoxy]pyrimidin-4-yl]-2,8-diazaspiro[4.5]decane-3-carboxylic acid | | Descriptor: | (3~{S})-8-[2-azanyl-6-[(1~{R})-1-(4-chloranyl-2-phenyl-phenyl)-2,2,2-tris(fluoranyl)ethoxy]pyrimidin-4-yl]-2,8-diazaspiro[4.5]decane-3-carboxylic acid, FE (III) ION, Tryptophan 5-hydroxylase 1 | | Authors: | Stein, A.J, Goldberg, D.R, De Lombaert, S, Holt, M.C. | | Deposit date: | 2016-07-26 | | Release date: | 2017-01-25 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Optimization of spirocyclic proline tryptophan hydroxylase-1 inhibitors.
Bioorg. Med. Chem. Lett., 27, 2017
|
|
5TPG
 
 | | Optimization of spirocyclic proline tryptophanhydroxylase-1 inhibitors | | Descriptor: | (3S)-8-(2-amino-6-{(1R)-1-[5-chloro-3'-(methylsulfonyl)[1,1'-biphenyl]-2-yl]-2,2,2-trifluoroethoxy}pyrimidin-4-yl)-2,8-diazaspiro[4.5]decane-3-carboxylic acid, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ACETONITRILE, ... | | Authors: | Stein, A.J, Goldberg, D.R, De Lombaert, S, Holt, M.C. | | Deposit date: | 2016-10-20 | | Release date: | 2017-01-25 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Optimization of spirocyclic proline tryptophan hydroxylase-1 inhibitors.
Bioorg. Med. Chem. Lett., 27, 2017
|
|
5PAH
 
 | |
6PAH
 
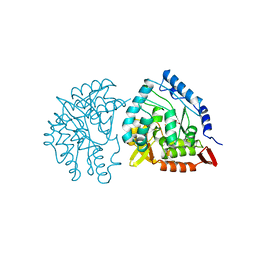 | | HUMAN PHENYLALANINE HYDROXYLASE CATALYTIC DOMAIN DIMER WITH BOUND L-DOPA (3,4-DIHYDROXYPHENYLALANINE) INHIBITOR | | Descriptor: | 3,4-DIHYDROXYPHENYLALANINE, FE (III) ION, PHENYLALANINE 4-MONOOXYGENASE | | Authors: | Erlandsen, H, Flatmark, T, Stevens, R.C. | | Deposit date: | 1998-08-20 | | Release date: | 1999-04-27 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystallographic analysis of the human phenylalanine hydroxylase catalytic domain with bound catechol inhibitors at 2.0 A resolution.
Biochemistry, 37, 1998
|
|
7VGM
 
 | |
3PAH
 
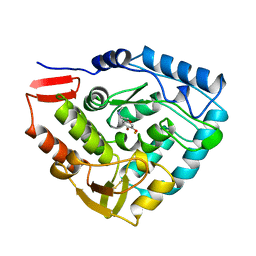 | | HUMAN PHENYLALANINE HYDROXYLASE CATALYTIC DOMAIN DIMER WITH BOUND ADRENALINE INHIBITOR | | Descriptor: | 4-[(1S)-1-hydroxy-2-(methylamino)ethyl]benzene-1,2-diol, FE (III) ION, PHENYLALANINE HYDROXYLASE | | Authors: | Erlandsen, H, Flatmark, T, Stevens, R.C. | | Deposit date: | 1998-08-20 | | Release date: | 1999-04-27 | | Last modified: | 2013-03-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystallographic analysis of the human phenylalanine hydroxylase catalytic domain with bound catechol inhibitors at 2.0 A resolution.
Biochemistry, 37, 1998
|
|
3E2T
 
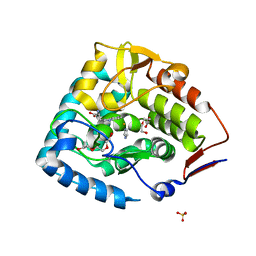 | | The catalytic domain of chicken tryptophan hydroxylase 1 with bound tryptophan | | Descriptor: | FE (III) ION, IMIDAZOLE, SULFATE ION, ... | | Authors: | Windahl, M.S, Petersen, C.R, Christensen, H.E.C, Harris, P. | | Deposit date: | 2008-08-06 | | Release date: | 2008-11-04 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of tryptophan hydroxylase with bound amino acid substrate
Biochemistry, 47, 2008
|
|
1PAH
 
 | |
4PAH
 
 | |
4JPY
 
 | |
4JPX
 
 | |
4Q3Y
 
 | |
2V28
 
 | | Apo structure of the cold active phenylalanine hydroxylase from Colwellia psychrerythraea 34H | | Descriptor: | PHENYLALANINE-4-HYDROXYLASE, SULFATE ION | | Authors: | Leiros, H.-K.S, Pey, A.L, Innselset, M, Moe, E, Leiros, I, Steen, I.H, Martinez, A. | | Deposit date: | 2007-06-04 | | Release date: | 2007-06-19 | | Last modified: | 2018-10-24 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure of Phenylalanine Hydroxylase from Colwellia Psychrerythraea 34H, a Monomeric Cold Active Enzyme with Local Flexibility Around the Active Site and High Overall Stability.
J.Biol.Chem., 282, 2007
|
|
4Q3Z
 
 | |
2V27
 
 | | Structure of the cold active phenylalanine hydroxylase from Colwellia psychrerythraea 34H | | Descriptor: | FE (III) ION, PHENYLALANINE HYDROXYLASE, SULFATE ION | | Authors: | Leiros, H.-K.S, Pey, A.L, Innselset, M, Moe, E, Leiros, I, Steen, I.H, Martinez, A. | | Deposit date: | 2007-06-03 | | Release date: | 2007-06-19 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure of Phenylalanine Hydroxylase from Colwellia Psychrerythraea 34H, a Monomeric Cold Active Enzyme with Local Flexibility Around the Active Site and High Overall Stability.
J.Biol.Chem., 282, 2007
|
|
1TG2
 
 | | Crystal structure of phenylalanine hydroxylase A313T mutant with 7,8-dihydrobiopterin bound | | Descriptor: | 2-AMINO-6-(1,2-DIHYDROXY-PROPYL)-7,8-DIHYDRO-6H-PTERIDIN-4-ONE, FE (III) ION, Phenylalanine-4-hydroxylase | | Authors: | Erlandsen, H, Pey, A.L, Gamez, A, Perez, B, Desviat, L.R, Aguado, C, Koch, R, Surendran, S, Tyring, S, Matalon, R, Scriver, C.R, Ugarte, M, Martinez, A, Stevens, R.C. | | Deposit date: | 2004-05-28 | | Release date: | 2004-11-30 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Correction of kinetic and stability defects by tetrahydrobiopterin in phenylketonuria patients with certain phenylalanine hydroxylase mutations.
Proc.Natl.Acad.Sci.Usa, 101, 2004
|
|
4Q3W
 
 | | Crystal structure of C. violaceum phenylalanine hydroxylase D139E mutation | | Descriptor: | 1,2-ETHANEDIOL, COBALT (II) ION, Phenylalanine-4-hydroxylase | | Authors: | Ronau, J.A, Abu-Omar, M.M, Das, C. | | Deposit date: | 2014-04-12 | | Release date: | 2015-02-18 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | A conserved acidic residue in phenylalanine hydroxylase contributes to cofactor affinity and catalysis.
Biochemistry, 53, 2014
|
|
4Q3X
 
 | |
1TDW
 
 | | Crystal structure of double truncated human phenylalanine hydroxylase BH4-responsive PKU mutant A313T. | | Descriptor: | FE (III) ION, Phenylalanine-4-hydroxylase | | Authors: | Erlandsen, H, Pey, A.L, Gamez, A, Perez, B, Desviat, L.R, Aguado, C, Koch, R, Surendran, S, Tyring, S, Matalon, R, Scriver, C.R, Ugarte, M, Martinez, A, Stevens, R.C. | | Deposit date: | 2004-05-24 | | Release date: | 2004-11-30 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Correction of kinetic and stability defects by tetrahydrobiopterin in phenylketonuria patients with certain phenylalanine hydroxylase mutations.
Proc.Natl.Acad.Sci.Usa, 101, 2004
|
|
1TOH
 
 | |
3HFB
 
 | |
3HF6
 
 | | Crystal structure of human tryptophan hydroxylase type 1 with bound LP-521834 and FE | | Descriptor: | 4-(4-amino-6-{[(1R)-1-naphthalen-2-ylethyl]amino}-1,3,5-triazin-2-yl)-L-phenylalanine, FE (III) ION, Tryptophan 5-hydroxylase 1 | | Authors: | Tari, L.W, Swanson, R.V, Hunter, M.J. | | Deposit date: | 2009-05-11 | | Release date: | 2009-11-24 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Mechanism of Inhibition of Novel Tryptophan Hydroxylase Inhibitors Revealed by Co-crystal Structures and Kinetic Analysis
Curr Chem Genomics, 4, 2010
|
|
3HF8
 
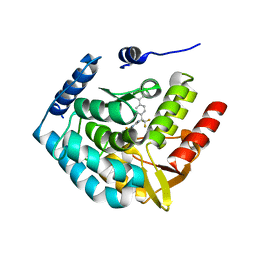 | | Crystal structure of human tryoptophan hydroxylase type 1 with bound LP-533401 and Fe | | Descriptor: | 4-{2-amino-6-[(1R)-2,2,2-trifluoro-1-(3'-fluorobiphenyl-4-yl)ethoxy]pyrimidin-4-yl}-L-phenylalanine, FE (III) ION, Tryptophan 5-hydroxylase 1 | | Authors: | Tari, L.W, Swanson, R.V, Hunter, M.J. | | Deposit date: | 2009-05-11 | | Release date: | 2010-04-21 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Mechanism of Inhibition of Novel Tryptophan Hydroxylase Inhibitors Revealed by Co-crystal Structures and Kinetic Analysis.
Curr Chem Genomics, 4, 2010
|
|
