8J48
 
 | | Crystal structure of OY phytoplasma SAP05 in complex with AtGATA18 | | Descriptor: | GATA transcription factor 18, Sequence-variable mosaic (SVM) signal sequence domain-containing protein, ZINC ION | | Authors: | Dong, C, Yan, X, Yuan, X. | | Deposit date: | 2023-04-19 | | Release date: | 2024-02-28 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Molecular basis of SAP05-mediated ubiquitin-independent proteasomal degradation of transcription factors
Nat Commun, 15, 2024
|
|
7AOA
 
 | | Structure of the extended MTA1/HDAC1/MBD2/RBBP4 NURD deacetylase complex | | Descriptor: | Histone deacetylase 1, Histone-binding protein RBBP4, INOSITOL HEXAKISPHOSPHATE, ... | | Authors: | Millard, C.J, Fairall, L, Ragan, T.J, Savva, C.G, Schwabe, J.W.R. | | Deposit date: | 2020-10-14 | | Release date: | 2020-11-11 | | Last modified: | 2020-12-30 | | Method: | ELECTRON MICROSCOPY (19.4 Å) | | Cite: | The topology of chromatin-binding domains in the NuRD deacetylase complex.
Nucleic Acids Res., 48, 2020
|
|
7AO9
 
 | | Structure of the core MTA1/HDAC1/MBD2 NURD deacetylase complex | | Descriptor: | Histone deacetylase 1, INOSITOL HEXAKISPHOSPHATE, Metastasis-associated protein MTA1, ... | | Authors: | Millard, C.J, Fairall, L, Ragan, T.J, Savva, C.G, Schwabe, J.W.R. | | Deposit date: | 2020-10-14 | | Release date: | 2020-11-11 | | Last modified: | 2020-12-30 | | Method: | ELECTRON MICROSCOPY (6.1 Å) | | Cite: | The topology of chromatin-binding domains in the NuRD deacetylase complex.
Nucleic Acids Res., 48, 2020
|
|
7AO8
 
 | | Structure of the MTA1/HDAC1/MBD2 NURD deacetylase complex | | Descriptor: | Histone deacetylase 1, INOSITOL HEXAKISPHOSPHATE, Metastasis-associated protein MTA1, ... | | Authors: | Millard, C.J, Fairall, L, Ragan, T.J, Savva, C.G, Schwabe, J.W.R. | | Deposit date: | 2020-10-14 | | Release date: | 2020-11-11 | | Last modified: | 2020-12-30 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | The topology of chromatin-binding domains in the NuRD deacetylase complex.
Nucleic Acids Res., 48, 2020
|
|
6ZFV
 
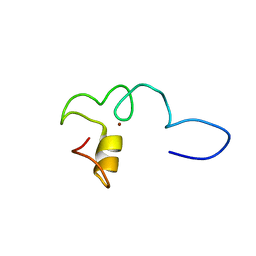 | |
5O9B
 
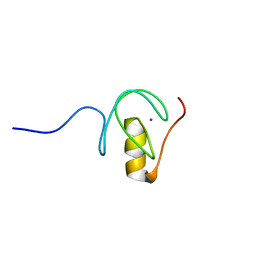 | |
2M9W
 
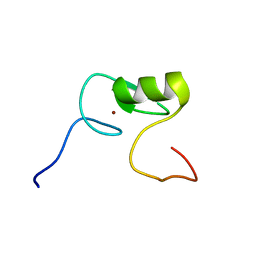 | | Solution NMR Structure of Transcription Factor GATA-4 from Homo sapiens, Northeast Structural Genomics Consortium (NESG) Target HR4783B | | Descriptor: | Transcription factor GATA-4, ZINC ION | | Authors: | Xu, X, Eletsky, A, Lee, D, Kohn, E, Janjua, H, Xiao, R, Acton, T.B, Everett, J.K, Montelione, G.T, Szyperski, T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2013-06-20 | | Release date: | 2013-07-10 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Solution NMR Structure of a Transcription Factor GATA-4 from Homo sapiens, Northeast Structural Genomics Consortium (NESG) Target HR4783B
To be Published
|
|
4HCA
 
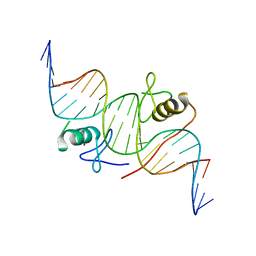 | | DNA binding by GATA transcription factor-complex 1 | | Descriptor: | DNA (5'-D(*AP*AP*TP*GP*TP*CP*CP*AP*TP*CP*TP*GP*AP*TP*AP*AP*GP*AP*CP*G)-3'), DNA (5'-D(*TP*TP*CP*GP*TP*CP*TP*TP*AP*TP*CP*AP*GP*AP*TP*GP*GP*AP*CP*A)-3'), Trans-acting T-cell-specific transcription factor GATA-3, ... | | Authors: | Chen, Y, Bates, D.L, Dey, R, Chen, L. | | Deposit date: | 2012-09-28 | | Release date: | 2012-12-05 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | DNA Binding by GATA Transcription Factor Suggests Mechanisms of DNA Looping and Long-Range Gene Regulation.
Cell Rep, 2, 2012
|
|
4HC7
 
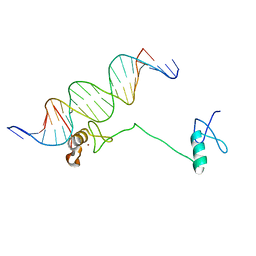 | | Crystal structure of the full DNA binding domain of GATA3-complex 2 | | Descriptor: | DNA (5'-D(*AP*AP*GP*GP*TP*TP*AP*TP*CP*TP*CP*TP*GP*AP*TP*TP*TP*AP*GP*G)-3'), DNA (5'-D(*TP*TP*CP*CP*TP*AP*AP*AP*TP*CP*AP*GP*AP*GP*AP*TP*AP*AP*CP*C)-3'), Trans-acting T-cell-specific transcription factor GATA-3, ... | | Authors: | Chen, Y, Bates, D.L, Dey, R, Chen, L. | | Deposit date: | 2012-09-28 | | Release date: | 2012-12-05 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | DNA Binding by GATA Transcription Factor Suggests Mechanisms of DNA Looping and Long-Range Gene Regulation.
Cell Rep, 2, 2012
|
|
4HC9
 
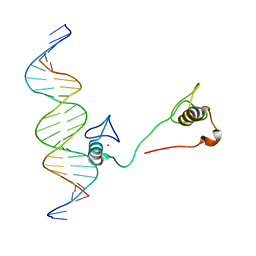 | | DNA binding by GATA transcription factor-complex 3 | | Descriptor: | DNA (5'-D(*AP*AP*GP*GP*TP*TP*AP*TP*CP*TP*CP*TP*GP*AP*TP*TP*TP*AP*GP*G)-3'), DNA (5'-D(*TP*TP*CP*CP*TP*AP*AP*AP*TP*CP*AP*GP*AP*GP*AP*TP*AP*AP*CP*C)-3'), Trans-acting T-cell-specific transcription factor GATA-3, ... | | Authors: | Chen, Y, Bates, D.L, Dey, R, Chen, L. | | Deposit date: | 2012-09-28 | | Release date: | 2012-12-05 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | DNA Binding by GATA Transcription Factor Suggests Mechanisms of DNA Looping and Long-Range Gene Regulation.
Cell Rep, 2, 2012
|
|
3VEK
 
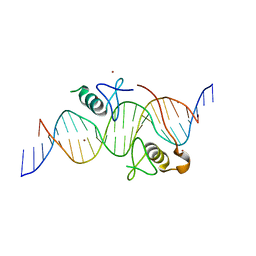 | | Both Zn Fingers of GATA1 Bound to Palindromic DNA Recognition Site, P1 Crystal Form | | Descriptor: | DNA (5'-D(*AP*AP*GP*AP*GP*TP*CP*CP*AP*TP*CP*TP*GP*AP*TP*AP*AP*GP*AP*C)-3'), DNA (5'-D(*TP*TP*GP*TP*CP*TP*TP*AP*TP*CP*AP*GP*AP*TP*GP*GP*AP*CP*TP*C)-3'), Erythroid transcription factor, ... | | Authors: | Jacques, D.A, Ripin, N, Wilkinson-White, L.E, Guss, J.M, Matthews, J.M. | | Deposit date: | 2012-01-09 | | Release date: | 2013-01-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.63 Å) | | Cite: | GATA1 directly mediates interactions with closely spaced pseudopalindromic but not distantly spaced double GATA sites on DNA.
Protein Sci., 24, 2015
|
|
3VD6
 
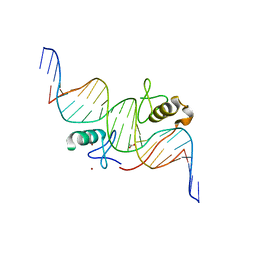 | | Both Zn Fingers of GATA1 Bound to Palindromic DNA Recognition Site, P21 Crystal Form | | Descriptor: | ACETATE ION, DNA (5'-D(*AP*AP*GP*AP*GP*TP*CP*CP*AP*TP*CP*TP*GP*AP*TP*AP*AP*GP*AP*C)-3'), DNA (5'-D(*TP*TP*GP*TP*CP*TP*TP*AP*TP*CP*AP*GP*AP*TP*GP*GP*AP*CP*TP*C)-3'), ... | | Authors: | Jacques, D.A, Ripin, N, Wilkinson-White, L.E, Guss, J.M, Matthews, J.M. | | Deposit date: | 2012-01-04 | | Release date: | 2013-01-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | GATA1 directly mediates interactions with closely spaced pseudopalindromic but not distantly spaced double GATA sites on DNA.
Protein Sci., 24, 2015
|
|
2L6Z
 
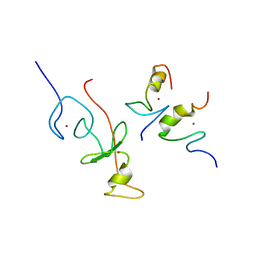 | | haddock model of GATA1NF:Lmo2LIM2-Ldb1LID with FOG | | Descriptor: | Erythroid transcription factor, LIM domain only 2, linker, ... | | Authors: | Wilkinson-White, L, Gamsjaeger, R, Dastmalchi, S, Wienert, B, Stokes, P.H, Crossley, M, Mackay, J.P, Matthews, J.M. | | Deposit date: | 2010-12-01 | | Release date: | 2011-08-31 | | Last modified: | 2011-09-28 | | Method: | SOLUTION NMR | | Cite: | Structural basis of simultaneous recruitment of the transcriptional regulators LMO2 and FOG1/ZFPM1 by the transcription factor GATA1
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
2L6Y
 
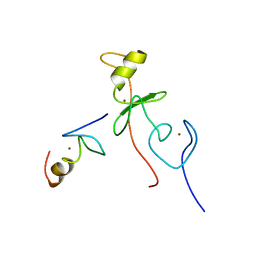 | | haddock model of GATA1NF:Lmo2LIM2-Ldb1LID | | Descriptor: | Erythroid transcription factor, LIM domain only 2, linker, ... | | Authors: | Wilkinson-White, L, Gamsjaeger, R, Dastmalchi, S, Wienert, B, Stokes, P.H, Crossley, M, Mackay, J.P, Matthews, J.M. | | Deposit date: | 2010-12-01 | | Release date: | 2011-08-31 | | Last modified: | 2011-09-28 | | Method: | SOLUTION NMR | | Cite: | Structural basis of simultaneous recruitment of the transcriptional regulators LMO2 and FOG1/ZFPM1 by the transcription factor GATA1
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
3DFV
 
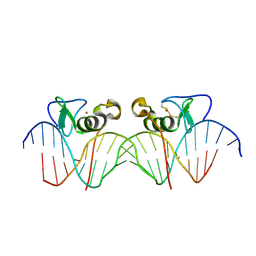 | | Adjacent GATA DNA binding | | Descriptor: | DNA (5'-D(*DAP*DAP*DGP*DCP*DAP*DGP*DAP*DTP*DAP*DAP*DGP*DTP*DCP*DTP*DTP*DAP*DTP*DCP*DAP*DG)-3'), DNA (5'-D(*DTP*DTP*DCP*DTP*DGP*DAP*DTP*DAP*DAP*DGP*DAP*DCP*DTP*DTP*DAP*DTP*DCP*DTP*DGP*DC)-3'), Trans-acting T-cell-specific transcription factor GATA-3, ... | | Authors: | Bates, D.L, Kim, G.K, Guo, L, Chen, L. | | Deposit date: | 2008-06-12 | | Release date: | 2008-07-29 | | Last modified: | 2017-10-25 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Crystal structures of multiple GATA zinc fingers bound to DNA reveal new insights into DNA recognition and self-association by GATA.
J.Mol.Biol., 381, 2008
|
|
3DFX
 
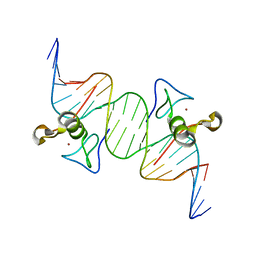 | | Opposite GATA DNA binding | | Descriptor: | DNA (5'-D(*DAP*DAP*DGP*DGP*DTP*DTP*DAP*DTP*DCP*DTP*DCP*DTP*DGP*DAP*DTP*DTP*DTP*DAP*DTP*DC)-3'), DNA (5'-D(*DTP*DTP*DGP*DAP*DTP*DAP*DAP*DAP*DTP*DCP*DAP*DGP*DAP*DGP*DAP*DTP*DAP*DAP*DCP*DC)-3'), Trans-acting T-cell-specific transcription factor GATA-3, ... | | Authors: | Bates, D.L, Kim, G.K, Guo, L, Chen, L. | | Deposit date: | 2008-06-12 | | Release date: | 2008-07-29 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structures of multiple GATA zinc fingers bound to DNA reveal new insights into DNA recognition and self-association by GATA.
J.Mol.Biol., 381, 2008
|
|
2VUT
 
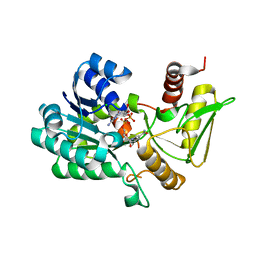 | | Crystal structure of NAD-bound NmrA-AreA zinc finger complex | | Descriptor: | CHLORIDE ION, GLYCEROL, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Kotaka, M, Johnson, C, Lamb, H.K, Hawkins, A.R, Ren, J, Stammers, D.K. | | Deposit date: | 2008-05-30 | | Release date: | 2008-07-29 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural Analysis of the Recognition of the Negative Regulator Nmra and DNA by the Zinc Finger from the Gata-Type Transcription Factor Area.
J.Mol.Biol., 381, 2008
|
|
2VUU
 
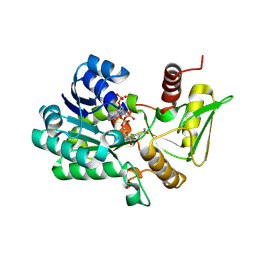 | | Crystal structure of NADP-bound NmrA-AreA zinc finger complex | | Descriptor: | NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, NITROGEN METABOLITE REPRESSION REGULATOR NMRA, NITROGEN REGULATORY PROTEIN AREA, ... | | Authors: | Kotaka, M, Johnson, C, Lamb, H.K, Hawkins, A.R, Ren, J, Stammers, D.K. | | Deposit date: | 2008-05-30 | | Release date: | 2008-07-29 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural Analysis of the Recognition of the Negative Regulator Nmra and DNA by the Zinc Finger from the Gata-Type Transcription Factor Area.
J.Mol.Biol., 381, 2008
|
|
2VUS
 
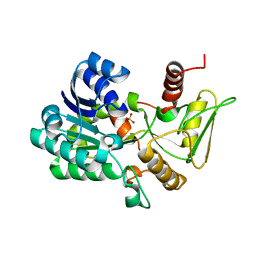 | | Crystal structure of unliganded NmrA-AreA zinc finger complex | | Descriptor: | CHLORIDE ION, NITROGEN METABOLITE REPRESSION REGULATOR NMRA, NITROGEN REGULATORY PROTEIN AREA, ... | | Authors: | Kotaka, M, Johnson, C, Lamb, H.K, Hawkins, A.R, Ren, J, Stammers, D.K. | | Deposit date: | 2008-05-30 | | Release date: | 2008-07-29 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural Analysis of the Recognition of the Negative Regulator Nmra and DNA by the Zinc Finger from the Gata-Type Transcription Factor Area.
J.Mol.Biol., 381, 2008
|
|
1Y0J
 
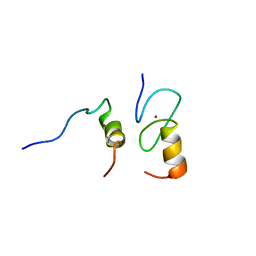 | | Zinc fingers as protein recognition motifs: structural basis for the GATA-1/Friend of GATA interaction | | Descriptor: | Erythroid transcription factor, ZINC ION, Zinc-finger protein ush | | Authors: | Liew, C.K, Simpson, R.J.Y, Kwan, A.H.Y, Crofts, L.A, Loughlin, F.E, Matthews, J.M, Crossley, M, Mackay, J.P. | | Deposit date: | 2004-11-15 | | Release date: | 2005-01-25 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | Zinc fingers as protein recognition motifs: Structural basis for the GATA-1/Friend of GATA interaction
Proc.Natl.Acad.Sci.Usa, 102, 2005
|
|
1GNF
 
 | | SOLUTION STRUCTURE OF THE N-TERMINAL ZINC FINGER OF MURINE GATA-1, NMR, 25 STRUCTURES | | Descriptor: | TRANSCRIPTION FACTOR GATA-1, ZINC ION | | Authors: | Kowalski, K, Czolij, R, King, G.F, Crossley, M, Mackay, J.P. | | Deposit date: | 1998-10-12 | | Release date: | 1999-06-08 | | Last modified: | 2017-11-29 | | Method: | SOLUTION NMR | | Cite: | The solution structure of the N-terminal zinc finger of GATA-1 reveals a specific binding face for the transcriptional co-factor FOG.
J.Biomol.NMR, 13, 1999
|
|
2GAT
 
 | | SOLUTION STRUCTURE OF THE C-TERMINAL DOMAIN OF CHICKEN GATA-1 BOUND TO DNA, NMR, REGULARIZED MEAN STRUCTURE | | Descriptor: | DNA (5'-D(*AP*AP*TP*GP*TP*TP*TP*AP*TP*CP*TP*GP*CP*AP*AP*C)-3'), DNA (5'-D(*GP*TP*TP*GP*CP*AP*GP*AP*TP*AP*AP*AP*CP*AP*TP*T)-3'), ERYTHROID TRANSCRIPTION FACTOR GATA-1, ... | | Authors: | Clore, G.M, Tjandra, N, Starich, M, Omichinski, J.G, Gronenborn, A.M. | | Deposit date: | 1997-11-07 | | Release date: | 1998-01-28 | | Last modified: | 2022-03-09 | | Method: | SOLUTION NMR | | Cite: | Use of dipolar 1H-15N and 1H-13C couplings in the structure determination of magnetically oriented macromolecules in solution.
Nat.Struct.Biol., 4, 1997
|
|
4GAT
 
 | | SOLUTION NMR STRUCTURE OF THE WILD TYPE DNA BINDING DOMAIN OF AREA COMPLEXED TO A 13BP DNA CONTAINING A CGATA SITE, REGULARIZED MEAN STRUCTURE | | Descriptor: | DNA (5'-D(*CP*AP*GP*CP*GP*AP*TP*AP*GP*AP*GP*AP*C)-3'), DNA (5'-D(*GP*TP*CP*TP*CP*TP*AP*TP*CP*GP*CP*TP*G)-3'), NITROGEN REGULATORY PROTEIN AREA, ... | | Authors: | Clore, G.M, Starich, M, Wikstrom, M, Gronenborn, A.M. | | Deposit date: | 1997-11-07 | | Release date: | 1998-01-28 | | Last modified: | 2022-03-16 | | Method: | SOLUTION NMR | | Cite: | The solution structure of a fungal AREA protein-DNA complex: an alternative binding mode for the basic carboxyl tail of GATA factors.
J.Mol.Biol., 277, 1998
|
|
7GAT
 
 | | SOLUTION NMR STRUCTURE OF THE L22V MUTANT DNA BINDING DOMAIN OF AREA COMPLEXED TO A 13 BP DNA CONTAINING A TGATA SITE, 34 STRUCTURES | | Descriptor: | DNA (5'-D(*CP*AP*GP*TP*GP*AP*TP*AP*GP*AP*GP*AP*C)-3'), DNA (5'-D(*GP*TP*CP*TP*CP*TP*AP*TP*CP*AP*CP*TP*G)-3'), NITROGEN REGULATORY PROTEIN AREA, ... | | Authors: | Clore, G.M, Starich, M, Wikstrom, M, Gronenborn, A.M. | | Deposit date: | 1997-11-07 | | Release date: | 1998-01-28 | | Last modified: | 2021-11-03 | | Method: | SOLUTION NMR | | Cite: | The solution structure of the Leu22-->Val mutant AREA DNA binding domain complexed with a TGATAG core element defines a role for hydrophobic packing in the determination of specificity.
J.Mol.Biol., 277, 1998
|
|
6GAT
 
 | | SOLUTION NMR STRUCTURE OF THE L22V MUTANT DNA BINDING DOMAIN OF AREA COMPLEXED TO A 13 BP DNA CONTAINING A TGATA SITE, REGULARIZED MEAN STRUCTURE | | Descriptor: | DNA (5'-D(*CP*AP*GP*TP*GP*AP*TP*AP*GP*AP*GP*AP*C)-3'), DNA (5'-D(*GP*TP*CP*TP*CP*TP*AP*TP*CP*AP*CP*TP*G)-3'), NITROGEN REGULATORY PROTEIN AREA, ... | | Authors: | Clore, G.M, Starich, M, Wikstrom, M, Gronenborn, A.M. | | Deposit date: | 1997-11-07 | | Release date: | 1998-01-28 | | Last modified: | 2021-11-03 | | Method: | SOLUTION NMR | | Cite: | The solution structure of the Leu22-->Val mutant AREA DNA binding domain complexed with a TGATAG core element defines a role for hydrophobic packing in the determination of specificity.
J.Mol.Biol., 277, 1998
|
|
