9B20
 
 | |
9B1Z
 
 | |
9B22
 
 | |
9B21
 
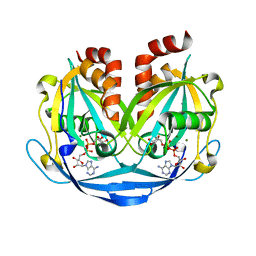 | | Crystal structure of ADP-ribose diphosphatase from Klebsiella pneumoniae (ADP Ribose bound, Orthorhombic P form) | | Descriptor: | ADP-ribose pyrophosphatase, MAGNESIUM ION, [(2R,3S,4R,5R)-5-(6-AMINOPURIN-9-YL)-3,4-DIHYDROXY-OXOLAN-2-YL]METHYL [HYDROXY-[[(2R,3S,4R,5S)-3,4,5-TRIHYDROXYOXOLAN-2-YL]METHOXY]PHOSPHORYL] HYDROGEN PHOSPHATE | | Authors: | Seattle Structural Genomics Center for Infectious Disease, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2024-03-14 | | Release date: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of ADP-ribose diphosphatase from Klebsiella pneumoniae (ADP Ribose bound, Orthorhombic P form)
To be published
|
|
8G9C
 
 | | Diphosphoinositol polyphosphate phosphohydrolase 1 (DIPP1/NUDT3) in complex with 5- difluoromethylenebisphosphonate inositol pentakisphosphate (5-PCF2P-IP5), an analogue of 5-InsP7 | | Descriptor: | CHLORIDE ION, Diphosphoinositol polyphosphate phosphohydrolase 1, FLUORIDE ION, ... | | Authors: | Zong, G, Wang, H, Shears, S. | | Deposit date: | 2023-02-21 | | Release date: | 2024-01-03 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Fluorination Influences the Bioisostery of Myo-Inositol Pyrophosphate Analogs.
Chemistry, 29, 2023
|
|
8G9D
 
 | | Diphosphoinositol polyphosphate phosphohydrolase 1 (DIPP1/NUDT3) in complex with 5- phosphonodifluoroacetamide inositol pentakisphosphate (5-PCF2Am-InsP5), an analogue of 5-InsP7 | | Descriptor: | (1,1-difluoro-2-oxo-2-{[(1s,2R,3S,4s,5R,6S)-2,3,4,5,6-pentakis(phosphonooxy)cyclohexyl]amino}ethyl)phosphonic acid, Diphosphoinositol polyphosphate phosphohydrolase 1 | | Authors: | Zong, G, Wang, H, Shears, S. | | Deposit date: | 2023-02-21 | | Release date: | 2024-01-03 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Fluorination Influences the Bioisostery of Myo-Inositol Pyrophosphate Analogs.
Chemistry, 29, 2023
|
|
7R0D
 
 | |
8SXS
 
 | |
8TFA
 
 | | Diphosphoinositol polyphosphate phosphohydrolase 1 (DIPP1/NUDT3) in complex with myo-5-PP-IP4, produced upon myo-(2OH)-IP5 phosphorylation by TvIPK | | Descriptor: | (1r,2R,3S,4r,5R,6S)-4-hydroxy-2,3,5,6-tetrakis(phosphonooxy)cyclohexyl trihydrogen diphosphate, CHLORIDE ION, Diphosphoinositol polyphosphate phosphohydrolase 1, ... | | Authors: | Zong, G, Wang, H, Shears, S.B. | | Deposit date: | 2023-07-09 | | Release date: | 2023-11-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Biochemical and structural characterization of an inositol pyrophosphate kinase from a giant virus.
Embo J., 43, 2024
|
|
8T98
 
 | | Diphosphoinositol polyphosphate phosphohydrolase 1 (DIPP1/NUDT3) in complex with Scyllo-3-PP-(1,2,4,5)IP4, Mg, and Fluoride ion | | Descriptor: | (1r,2R,3S,4r,5R,6S)-4-hydroxy-2,3,5,6-tetrakis(phosphonooxy)cyclohexyl trihydrogen diphosphate, CHLORIDE ION, Diphosphoinositol polyphosphate phosphohydrolase 1, ... | | Authors: | Zong, G, Wang, H, Shears, S.B. | | Deposit date: | 2023-06-23 | | Release date: | 2023-11-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Biochemical and structural characterization of an inositol pyrophosphate kinase from a giant virus.
Embo J., 43, 2024
|
|
8T99
 
 | | Diphosphoinositol polyphosphate phosphohydrolase 1 (DIPP1/NUDT3) in complex with Scyllo-L-1,4-[PP]2-(2,3)IP2, Mg, and Fluoride ion | | Descriptor: | (1R,2S,3S,4R,5S,6S)-2,3-dihydroxy-5,6-bis(phosphonooxy)cyclohexane-1,4-diyl bis[trihydrogen (diphosphate)], CHLORIDE ION, Diphosphoinositol polyphosphate phosphohydrolase 1, ... | | Authors: | Zong, G, Wang, H, Shears, S.B. | | Deposit date: | 2023-06-23 | | Release date: | 2023-11-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Biochemical and structural characterization of an inositol pyrophosphate kinase from a giant virus.
Embo J., 43, 2024
|
|
8TF9
 
 | | Diphosphoinositol polyphosphate phosphohydrolase 1 (DIPP1/NUDT3) in complex with myo-5-IP7, produced upon myo-IP6 phosphorylation by TvI | | Descriptor: | (1r,2R,3S,4s,5R,6S)-2,3,4,5,6-pentakis(phosphonooxy)cyclohexyl trihydrogen diphosphate, CHLORIDE ION, Diphosphoinositol polyphosphate phosphohydrolase 1, ... | | Authors: | Zong, G, Wang, H, Shears, S.B. | | Deposit date: | 2023-07-09 | | Release date: | 2023-11-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Biochemical and structural characterization of an inositol pyrophosphate kinase from a giant virus.
Embo J., 43, 2024
|
|
8A34
 
 | | MTH1 in complex with TH013071 | | Descriptor: | 5-(2-phenylphenyl)-1H-pyrimidine-2,4-dione, 7,8-dihydro-8-oxoguanine triphosphatase, ACETATE ION, ... | | Authors: | Liu, H, Scaletti, E, Stenmark, P. | | Deposit date: | 2022-06-07 | | Release date: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | MTH1 in complex with TH013071
To Be Published
|
|
8A0S
 
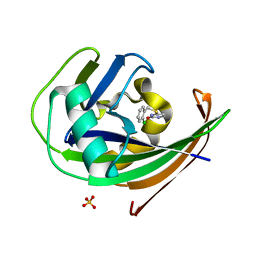 | | MTH1 in complex with TH013350 | | Descriptor: | 4-[(2-chlorophenyl)methylsulfanyl]-5~{H}-pyrimidin-2-one, 7,8-dihydro-8-oxoguanine triphosphatase, SULFATE ION | | Authors: | Liu, H, Scaletti, E, Stenmark, P. | | Deposit date: | 2022-05-30 | | Release date: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | MTH1 in complex with TH013350
To Be Published
|
|
8A3A
 
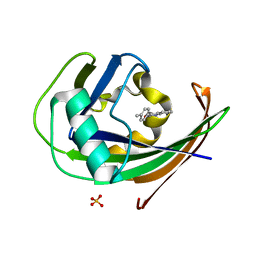 | | MTH1 in complex with TH013074 | | Descriptor: | 4-(4-methylphenyl)-1~{H}-quinazolin-2-one, 7,8-dihydro-8-oxoguanine triphosphatase, SULFATE ION | | Authors: | Liu, H, Scaletti, E, Stenmark, P. | | Deposit date: | 2022-06-07 | | Release date: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | MTH1 in complex with TH013074
To Be Published
|
|
8A07
 
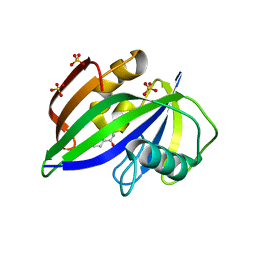 | |
8A0T
 
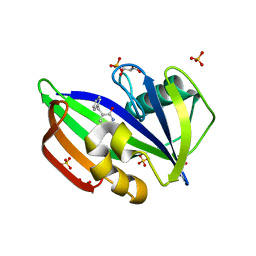 | | MTH1 in complex with TH012532 | | Descriptor: | 5-(phenylmethyl)pyrimidine-2,4-diol, 7,8-dihydro-8-oxoguanine triphosphatase, ACETATE ION, ... | | Authors: | Scaletti, E, Stenmark, P. | | Deposit date: | 2022-05-30 | | Release date: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | MTH1 in complex with TH012532
To Be Published
|
|
8I1G
 
 | |
8I18
 
 | |
8I19
 
 | |
8I8T
 
 | |
8I1E
 
 | | Crystal structure of human MTH1(G2K mutant) in complex with 2-oxo-dATP at pH 8.0 | | Descriptor: | 7,8-dihydro-8-oxoguanine triphosphatase, SODIUM ION, [[(2R,3S,5R)-5-(6-azanyl-2-oxidanylidene-1H-purin-9-yl)-3-oxidanyl-oxolan-2-yl]methoxy-oxidanyl-phosphoryl] phosphono hydrogen phosphate | | Authors: | Nakamura, T, Yamagata, Y. | | Deposit date: | 2023-01-13 | | Release date: | 2023-03-22 | | Last modified: | 2023-07-26 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Protonation states of Asp residues in the human Nudix hydrolase MTH1 contribute to its broad substrate recognition.
Febs Lett., 597, 2023
|
|
8I1J
 
 | | Crystal structure of human MTH1(G2K/D120N mutant) in complex with 2-oxo-dATP at pH 9.7 | | Descriptor: | 7,8-dihydro-8-oxoguanine triphosphatase, SODIUM ION, [[(2R,3S,5R)-5-(6-azanyl-2-oxidanylidene-1H-purin-9-yl)-3-oxidanyl-oxolan-2-yl]methoxy-oxidanyl-phosphoryl] phosphono hydrogen phosphate | | Authors: | Nakamura, T, Yamagata, Y. | | Deposit date: | 2023-01-13 | | Release date: | 2023-03-22 | | Last modified: | 2023-07-26 | | Method: | X-RAY DIFFRACTION (1.08 Å) | | Cite: | Protonation states of Asp residues in the human Nudix hydrolase MTH1 contribute to its broad substrate recognition.
Febs Lett., 597, 2023
|
|
8I1H
 
 | |
8I1I
 
 | | Crystal structure of human MTH1(G2K/D120N mutant) in complex with 2-oxo-dATP at pH 7.7 | | Descriptor: | 7,8-dihydro-8-oxoguanine triphosphatase, SODIUM ION, [[(2R,3S,5R)-5-(6-azanyl-2-oxidanylidene-1H-purin-9-yl)-3-oxidanyl-oxolan-2-yl]methoxy-oxidanyl-phosphoryl] phosphono hydrogen phosphate | | Authors: | Nakamura, T, Yamagata, Y. | | Deposit date: | 2023-01-13 | | Release date: | 2023-03-22 | | Last modified: | 2023-07-26 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Protonation states of Asp residues in the human Nudix hydrolase MTH1 contribute to its broad substrate recognition.
Febs Lett., 597, 2023
|
|
