3UCI
 
 | | Crystal structure of Rhodostomin ARLDDL mutant | | Descriptor: | disintegrin | | Authors: | Shiu, J.H, Chen, C.Y, Chen, Y.C, Chang, Y.T, Chang, Y.S, Huang, C.H, Chuang, W.J. | | Deposit date: | 2011-10-27 | | Release date: | 2012-11-21 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Design of Integrin AlphaVbeta3-Specific Disintegrin for Cancer Therapy
To be Published
|
|
1FVL
 
 | |
6LSQ
 
 | |
1Z1X
 
 | | Crystal Structure of a novel disintegrin from Saw-scaled viper at 3.2 A resolution | | Descriptor: | disintegrin | | Authors: | Hassan, M.I, Ethayathulla, A.S, Bilgrami, S, Singh, B, Yadav, S, Singh, T.P. | | Deposit date: | 2005-03-07 | | Release date: | 2005-04-19 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Crystal Structure of a novel disintegrin from Saw-scaled viper at 3.2A resolution
To be published
|
|
4R5R
 
 | |
4M4C
 
 | | Crystal structure of Rhodostomin ARGDP mutant | | Descriptor: | SULFATE ION, Zinc metalloproteinase/disintegrin | | Authors: | Chang, Y.T, Jeng, W.Y, Shiu, J.H, Chen, C.Y, Chuang, W.J. | | Deposit date: | 2013-08-07 | | Release date: | 2014-08-13 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Effect of C-terminal proline residue adjacent to the RGD motif in rhodostomin on its activity and structure
To be Published
|
|
8S9E
 
 | | Solution structure of jarastatin (rJast), a disintegrin from Bothrops jararaca | | Descriptor: | Disintegrin jarastatin | | Authors: | Vasconcelos, A.A, Estrada, J.E.C, Zingali, R.B, Almeida, F.C.L. | | Deposit date: | 2023-03-28 | | Release date: | 2023-11-29 | | Method: | SOLUTION NMR | | Cite: | Toward the mechanism of jarastatin (rJast) inhibition of the integrin alpha V beta 3
Int.J.Biol.Macromol., 255, 2024
|
|
4RQG
 
 | |
4R5U
 
 | |
1J2L
 
 | | Crystal structure of the disintegrin, trimestatin | | Descriptor: | Disintegrin triflavin, SULFATE ION | | Authors: | Fujii, Y, Okuda, D, Fujimoto, Z, Morita, T, Mizuno, H. | | Deposit date: | 2003-01-06 | | Release date: | 2003-10-07 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal Structure of Trimestatin, a Disintegrin Containing a Cell Adhesion Recognition Motif RGD
J.Mol.Biol., 332, 2003
|
|
3C05
 
 | | Crystal structure of Acostatin from Agkistrodon Contortrix Contortrix | | Descriptor: | Disintegrin acostatin alpha, Disintegrin acostatin-beta, SULFATE ION | | Authors: | Moiseeva, N, Bau, R, Allaire, M. | | Deposit date: | 2008-01-18 | | Release date: | 2008-04-29 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of acostatin, a dimeric disintegrin from southern copperhead
(Agkistrodon contortrix contortrix) at 1.7 A resolution
Acta Crystallogr.,Sect.D, 64, 2008
|
|
7X4Z
 
 | |
7X4V
 
 | |
7X4S
 
 | |
2PJI
 
 | |
2PJF
 
 | | Solution structure of rhodostomin | | Descriptor: | Rhodostoxin-disintegrin rhodostomin | | Authors: | Chuang, W.J, Chen, Y.C, Chen, C.Y, Chang, Y.T. | | Deposit date: | 2007-04-16 | | Release date: | 2007-05-08 | | Last modified: | 2022-03-16 | | Method: | SOLUTION NMR | | Cite: | Effect of D to E mutation of the RGD motif in rhodostomin on its activity, structure, and dynamics: Importance of the interactions between the D residue and integrin
Proteins, 2009
|
|
2PJG
 
 | | Solution structure of rhodostomin D51E mutant | | Descriptor: | Rhodostoxin-disintegrin rhodostomin | | Authors: | Chuang, W.J, Chen, Y.C, Chen, C.Y, Chou, L.J. | | Deposit date: | 2007-04-16 | | Release date: | 2007-05-08 | | Last modified: | 2021-11-10 | | Method: | SOLUTION NMR | | Cite: | Effect of D to E mutation of the RGD motif in rhodostomin on its activity, structure, and dynamics: Importance of the interactions between the D residue and integrin
Proteins, 2009
|
|
1L3X
 
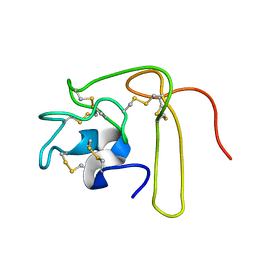 | | Solution Structure of Novel Disintegrin Salmosin | | Descriptor: | platelet aggregation inhibitor disintegrin | | Authors: | Shin, J, Lee, W. | | Deposit date: | 2002-03-01 | | Release date: | 2003-12-23 | | Last modified: | 2012-11-21 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a novel disintegrin, salmosin, from Agkistrondon halys venom
Biochemistry, 42, 2003
|
|
1N4Y
 
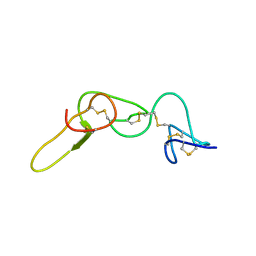 | | REFINED STRUCTURE OF KISTRIN | | Descriptor: | KISTRIN | | Authors: | Krezel, A.M, Krane, J, Dennis, M.S, Lazarus, R.A, Wagner, G. | | Deposit date: | 2002-11-02 | | Release date: | 2003-01-28 | | Last modified: | 2012-09-05 | | Method: | SOLUTION NMR | | Cite: | Solution structure of kistrin, a potent platelet aggregation inhibitor and GP IIb-IIIa antagonist.
Science, 253, 1991
|
|
1Q7J
 
 | |
1Q7I
 
 | |
1RMR
 
 | | Crystal Structure of Schistatin, a Disintegrin Homodimer from saw-scaled Viper (Echis carinatus) at 2.5 A resolution | | Descriptor: | Disintegrin schistatin | | Authors: | Bilgrami, S, Tomar, S, Yadav, S, Kaur, P, Kumar, J, Jabeen, T, Sharma, S, Singh, T.P. | | Deposit date: | 2003-11-28 | | Release date: | 2004-06-16 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of schistatin, a disintegrin homodimer from saw-scaled viper (Echis carinatus) at 2.5 A resolution
J.Mol.Biol., 341, 2004
|
|
1RO3
 
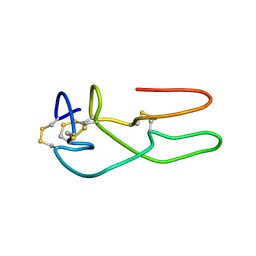 | | New structural insights on short disintegrin echistatin by NMR | | Descriptor: | Disintegrin echistatin | | Authors: | Monleon, D, Esteve, V, Calvete, J.J, Marcinkiewicz, C, Celda, B. | | Deposit date: | 2003-12-01 | | Release date: | 2003-12-09 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | Conformation and concerted dynamics of the integrin-binding site and the C-terminal region of echistatin revealed by homonuclear NMR
Biochem.J., 387, 2005
|
|
1TEJ
 
 | | Crystal structure of a disintegrin heterodimer at 1.9 A resolution. | | Descriptor: | disintegrin chain A, disintegrin chain B | | Authors: | Bilgrami, S, Kaur, P, Yadav, S, Perbandt, M, Betzel, C, Singh, T.P. | | Deposit date: | 2004-05-25 | | Release date: | 2004-06-15 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structure of the Disintegrin Heterodimer from Saw-Scaled Viper (Echis carinatus) at 1.9 A Resolution
Biochemistry, 44, 2005
|
|
2LJV
 
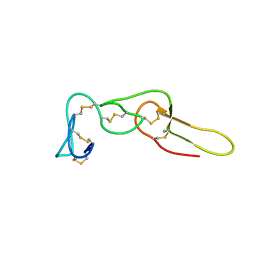 | | Solution structure of Rhodostomin G50L mutant | | Descriptor: | Disintegrin rhodostomin | | Authors: | Chuang, W, Shiu, J, Chen, C, Chen, Y, Chang, Y, Huang, C. | | Deposit date: | 2011-09-29 | | Release date: | 2012-10-03 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Design of Integrin AlphaVbeta3-Specific Disintegrin for Cancer Therapy
To be Published
|
|
