8U3B
 
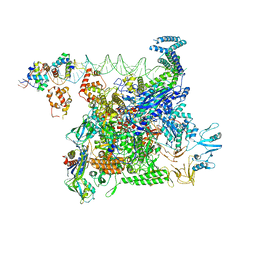 | | Cryo-EM structure of E. coli NarL-transcription activation complex at 3.2A | | Descriptor: | DNA (69-MER), DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, ... | | Authors: | Liu, B, Kompaniiets, D, Wang, D. | | Deposit date: | 2023-09-07 | | Release date: | 2024-01-17 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (3.23 Å) | | Cite: | Structural basis for transcription activation by the nitrate-responsive regulator NarL.
Nucleic Acids Res., 52, 2024
|
|
8A5R
 
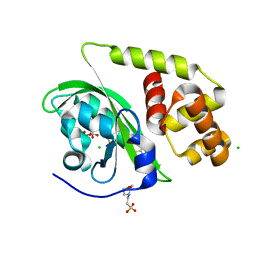 | | Crystal structure of light-activated DNA-binding protein EL222 from Erythrobacter litoralis crystallized and measured in dark. | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CHLORIDE ION, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Koval, T, Chaudhari, A, Fuertes, G, Andersson, I, Dohnalek, J. | | Deposit date: | 2022-06-15 | | Release date: | 2023-07-05 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | EL222 from Erythrobacter litoralis.
To Be Published
|
|
8A5S
 
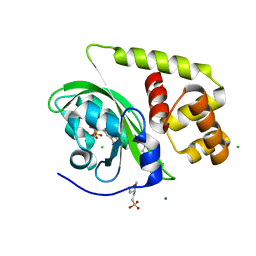 | | Crystal structure of light-activated DNA-binding protein EL222 from Erythrobacter litoralis crystallized in dark, measured illuminated. | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CHLORIDE ION, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Koval, T, Chaudhari, A, Fuertes, G, Andersson, I, Dohnalek, J. | | Deposit date: | 2022-06-15 | | Release date: | 2023-07-05 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | EL222 from Erythrobacter litoralis.
To Be Published
|
|
8B4A
 
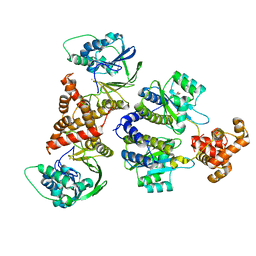 | | Nativ complex of PqsE and RhlR with autoinducer C4-HSL | | Descriptor: | 2-aminobenzoylacetyl-CoA thioesterase, FE (III) ION, N-[(3S)-2-oxotetrahydrofuran-3-yl]butanamide, ... | | Authors: | Borgert, S.R, Blankenfeldt, W. | | Deposit date: | 2022-09-20 | | Release date: | 2022-12-14 | | Method: | X-RAY DIFFRACTION (3.06 Å) | | Cite: | Moonlighting chaperone activity of the enzyme PqsE contributes to RhlR-controlled virulence of Pseudomonas aeruginosa.
Nat Commun, 13, 2022
|
|
7R3G
 
 | |
7R3I
 
 | |
7R3H
 
 | |
7R3J
 
 | | Nativ complex of PqsE and RhlR with the synthetic antagonist mBTL | | Descriptor: | 2-aminobenzoylacetyl-CoA thioesterase, 4-(3-bromophenoxy)-N-[(3S)-2-oxothiolan-3-yl]butanamide, FE (III) ION, ... | | Authors: | Borgert, S.R, Schmelz, S, Blankenfeldt, W. | | Deposit date: | 2022-02-07 | | Release date: | 2022-12-14 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.06 Å) | | Cite: | Moonlighting chaperone activity of the enzyme PqsE contributes to RhlR-controlled virulence of Pseudomonas aeruginosa.
Nat Commun, 13, 2022
|
|
7R3E
 
 | | Fusion construct of PqsE and RhlR in complex with the synthetic antagonist mBTL | | Descriptor: | 2-aminobenzoylacetyl-CoA thioesterase,Regulatory protein RhlR, 4-(3-bromophenoxy)-N-[(3S)-2-oxothiolan-3-yl]butanamide, FE (III) ION | | Authors: | Borgert, S.R, Schmelz, S, Blankenfeldt, W. | | Deposit date: | 2022-02-07 | | Release date: | 2022-12-14 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.46 Å) | | Cite: | Moonlighting chaperone activity of the enzyme PqsE contributes to RhlR-controlled virulence of Pseudomonas aeruginosa.
Nat Commun, 13, 2022
|
|
8DQ1
 
 | | Quorum-sensing receptor RhlR bound to PqsE | | Descriptor: | 2-aminobenzoylacetyl-CoA thioesterase, 4-(3-bromophenoxy)-N-[(3S)-2-oxothiolan-3-yl]butanamide, DNA (5'-D(*AP*CP*CP*TP*GP*CP*CP*AP*GP*AP*CP*TP*GP*CP*AP*CP*AP*G)-3'), ... | | Authors: | Paczkowski, J.E, Fromme, J.C, Feathers, J.R. | | Deposit date: | 2022-07-18 | | Release date: | 2022-12-07 | | Last modified: | 2022-12-14 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Structure of the RhlR-PqsE complex from Pseudomonas aeruginosa reveals mechanistic insights into quorum-sensing gene regulation.
Structure, 30, 2022
|
|
8DQ0
 
 | | Quorum-sensing receptor RhlR bound to PqsE | | Descriptor: | 2-aminobenzoylacetyl-CoA thioesterase, 4-(3-bromophenoxy)-N-[(3S)-2-oxothiolan-3-yl]butanamide, RhlR protein | | Authors: | Paczkowski, J.E, Fromme, J.C, Feathers, J.R. | | Deposit date: | 2022-07-18 | | Release date: | 2022-12-07 | | Last modified: | 2022-12-14 | | Method: | ELECTRON MICROSCOPY (3.74 Å) | | Cite: | Structure of the RhlR-PqsE complex from Pseudomonas aeruginosa reveals mechanistic insights into quorum-sensing gene regulation.
Structure, 30, 2022
|
|
7VIM
 
 | |
7PK5
 
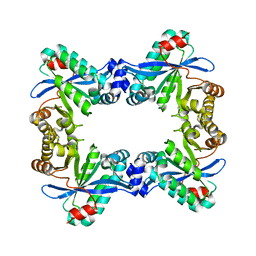 | |
7X1K
 
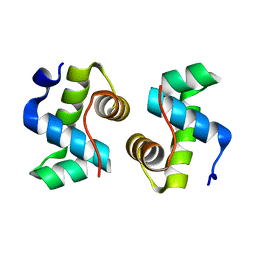 | |
7VE6
 
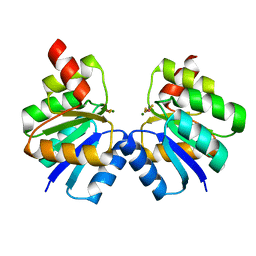 | | N-terminal domain of VraR | | Descriptor: | BERYLLIUM TRIFLUORIDE ION, MAGNESIUM ION, Response regulator protein VraR | | Authors: | Kumar, J.V, Chen, C, Hsu, C.H. | | Deposit date: | 2021-09-08 | | Release date: | 2022-05-18 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.77 Å) | | Cite: | Structural insights into DNA binding domain of vancomycin-resistance-associated response regulator in complex with its promoter DNA from Staphylococcus aureus.
Protein Sci., 31, 2022
|
|
7VE5
 
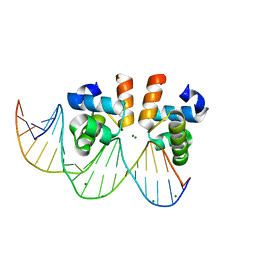 | | C-terminal domain of VraR | | Descriptor: | DNA-binding response regulator, MAGNESIUM ION, R1-DNA | | Authors: | Kumar, J.V, Chen, C, Hsu, C.H. | | Deposit date: | 2021-09-08 | | Release date: | 2022-05-18 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural insights into DNA binding domain of vancomycin-resistance-associated response regulator in complex with its promoter DNA from Staphylococcus aureus.
Protein Sci., 31, 2022
|
|
7DWM
 
 | | Crystal structure of the phage VqmA-DPO complex | | Descriptor: | 3,5-dimethylpyrazin-2-ol, Transcriptional regulator | | Authors: | Gu, Y, Yang, W.S. | | Deposit date: | 2021-01-17 | | Release date: | 2021-05-05 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Understanding the mechanism of asymmetric gene regulation determined by the VqmA of vibriophage.
Biochem.Biophys.Res.Commun., 558, 2021
|
|
6ZIX
 
 | | Structure of RcsB from Salmonella enterica serovar Typhimurium bound to promoter P1flhDC in the presence of phosphomimetic BeF3- | | Descriptor: | BERYLLIUM TRIFLUORIDE ION, MAGNESIUM ION, P1flhDC promoter sequence of 23 bp, ... | | Authors: | Huesa, J, Marina, A, Casino, P. | | Deposit date: | 2020-06-26 | | Release date: | 2021-02-17 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Structure-based analyses of Salmonella RcsB variants unravel new features of the Rcs regulon.
Nucleic Acids Res., 49, 2021
|
|
6ZJ2
 
 | | Structure of RcsB from Salmonella enterica serovar Typhimurium bound to promoter rprA in the presence of phosphomimetic BeF3- | | Descriptor: | BERYLLIUM TRIFLUORIDE ION, DNA (5'-D(P*CP*CP*GP*AP*TP*CP*AP*GP*AP*TP*TP*CP*GP*TP*CP*TP*CP*AP*AP*TP*AP*GP*G)-3'), MAGNESIUM ION, ... | | Authors: | Huesa, J, Marina, A, Casino, P. | | Deposit date: | 2020-06-27 | | Release date: | 2021-02-17 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.38 Å) | | Cite: | Structure-based analyses of Salmonella RcsB variants unravel new features of the Rcs regulon.
Nucleic Acids Res., 49, 2021
|
|
6V7X
 
 | | Structure of a phage-encoded quorum sensing anti-activator, Aqs1 bound to LasR | | Descriptor: | N-3-OXO-DODECANOYL-L-HOMOSERINE LACTONE, QUORUM SENSING ANTI-ACTIVATOR PROTEIN AQS1, Transcriptional regulator LasR | | Authors: | Shah, M, Moraes, T.F, Maxwell, K.L. | | Deposit date: | 2019-12-09 | | Release date: | 2020-12-16 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | A phage-encoded anti-activator inhibits quorum sensing in Pseudomonas aeruginosa.
Mol.Cell, 81, 2021
|
|
6V7W
 
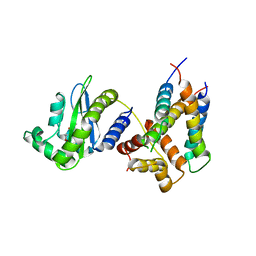 | |
6UGL
 
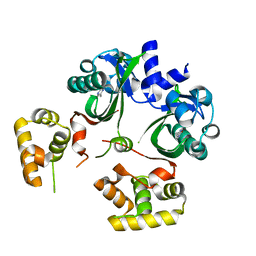 | | VqmA bound to DPO | | Descriptor: | 3,5-dimethylpyrazin-2(1H)-one, Helix-turn-helix transcriptional regulator | | Authors: | Paczkowski, J.E, Huang, X. | | Deposit date: | 2019-09-26 | | Release date: | 2020-01-29 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Mechanism underlying autoinducer recognition in theVibrio choleraeDPO-VqmA quorum-sensing pathway.
J.Biol.Chem., 295, 2020
|
|
6KJU
 
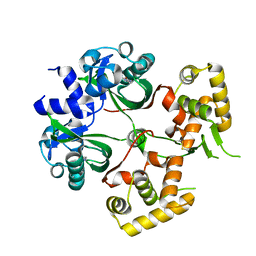 | | Huge conformation shift of Vibrio cholerae VqmA dimer in the absence of target DNA provides insight into DNA-binding mechanisms of LuxR-type receptors | | Descriptor: | 3,5-dimethylpyrazin-2-ol, Helix-turn-helix transcriptional regulator | | Authors: | Wu, H, Li, M.J, Guo, H.J, Zhou, H, Wang, W.W, Xu, Q, Xu, C.Y, Yu, F, He, J.H. | | Deposit date: | 2019-07-23 | | Release date: | 2019-11-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Large conformation shifts of Vibrio cholerae VqmA dimer in the absence of target DNA provide insight into DNA-binding mechanisms of LuxR-type receptors.
Biochem.Biophys.Res.Commun., 520, 2019
|
|
6JQS
 
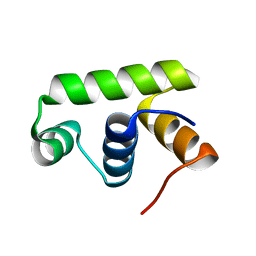 | | Structure of Transcription factor, GerE | | Descriptor: | DNA-binding response regulator | | Authors: | Lee, J.H, Lee, C.W. | | Deposit date: | 2019-04-01 | | Release date: | 2019-04-24 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Crystal structure of a transcription factor, GerE (PaGerE), from spore-forming bacterium Paenisporosarcina sp. TG-14.
Biochem.Biophys.Res.Commun., 513, 2019
|
|
6MWW
 
 | | LasR LBD:BB0126 complex | | Descriptor: | 4-[3-(methylsulfonyl)phenoxy]-N-[(1R,3R,5R)-2-oxobicyclo[3.1.0]hexan-3-yl]butanamide, Transcriptional regulator LasR | | Authors: | Bassler, B.L, Paczkowski, J.E. | | Deposit date: | 2018-10-30 | | Release date: | 2019-04-03 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.76 Å) | | Cite: | An Autoinducer Analogue Reveals an Alternative Mode of Ligand Binding for the LasR Quorum-Sensing Receptor.
Acs Chem.Biol., 14, 2019
|
|
