1Z6A
 
 | | Sulfolobus solfataricus SWI2/SNF2 ATPase core domain | | Descriptor: | Helicase of the snf2/rad54 family, MERCURY (II) ION, PHOSPHATE ION | | Authors: | Duerr, H, Koerner, C, Mueller, M, Hickmann, V, Hopfner, K.P. | | Deposit date: | 2005-03-22 | | Release date: | 2005-05-03 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | X-ray structures of the Sulfolobus solfataricus SWI2/SNF2 ATPase core and its complex with DNA
Cell(Cambridge,Mass.), 121, 2005
|
|
1Z63
 
 | | Sulfolobus solfataricus SWI2/SNF2 ATPase core in complex with dsDNA | | Descriptor: | 5'-D(*AP*AP*AP*AP*AP*A*AP*TP*TP*GP*CP*CP*GP*AP*AP*GP*AP*CP*GP*AP*AP*AP*AP*AP*A)-3', 5'-D(*TP*TP*TP*TP*TP*TP*TP*CP*GP*TP*CP*TP*TP*CP*GP*GP*CP*AP*AP*TP*TP*TP*TP*TP*T)-3', Helicase of the snf2/rad54 family | | Authors: | Duerr, H, Koerner, C, Mueller, M, Hickmann, V, Hopfner, K.P. | | Deposit date: | 2005-03-21 | | Release date: | 2005-05-17 | | Last modified: | 2017-08-23 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | X-ray structures of the Sulfolobus solfataricus SWI2/SNF2 ATPase core and its complex with DNA.
Cell(Cambridge,Mass.), 121, 2005
|
|
7XWY
 
 | |
7EPU
 
 | | Crystal structure of HsALC1 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Chromodomain-helicase-DNA-binding protein 1-like, MAGNESIUM ION, ... | | Authors: | Wang, L, Chen, K.J. | | Deposit date: | 2021-04-27 | | Release date: | 2021-07-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structural basis of ALC1/CHD1L autoinhibition and the mechanism of activation by the nucleosome.
Nat Commun, 12, 2021
|
|
3MWY
 
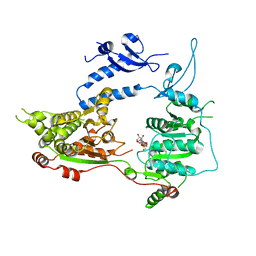 | |
4S20
 
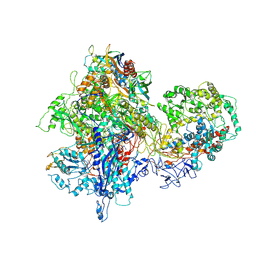 | | Structural basis for transcription reactivation by RapA | | Descriptor: | 5'-D(P*AP*CP*GP*AP*CP*TP*GP*AP*GP*CP*CP*GP*AP*TP*G)-3', 5'-R(P*AP*UP*CP*GP*GP*CP*UP*CP*A)-3', DNA-directed RNA polymerase subunit alpha, ... | | Authors: | Liu, B, Zuo, Y, Steitz, T.A. | | Deposit date: | 2015-01-16 | | Release date: | 2015-02-04 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (4.7 Å) | | Cite: | Structural basis for transcription reactivation by RapA.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
5GN1
 
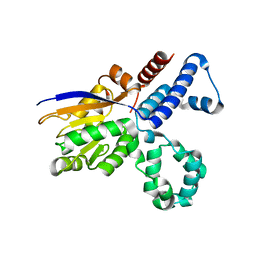 | |
5HZR
 
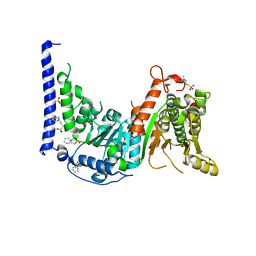 | | Crystal structure of MtSnf2 | | Descriptor: | 3-(1-methylpiperidinium-1-yl)propane-1-sulfonate, SNF2-family ATP dependent chromatin remodeling factor like protein, SODIUM ION, ... | | Authors: | Chen, Z.C, Xia, X. | | Deposit date: | 2016-02-02 | | Release date: | 2016-07-20 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Structure of chromatin remodeler Swi2/Snf2 in the resting state
Nat.Struct.Mol.Biol., 23, 2016
|
|
5JXR
 
 | | Crystal structure of MtISWI | | Descriptor: | CHLORIDE ION, Chromatin-remodeling complex ATPase-like protein | | Authors: | Chen, Z, Yan, L. | | Deposit date: | 2016-05-13 | | Release date: | 2016-12-07 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.404 Å) | | Cite: | Structure and regulation of the chromatin remodeller ISWI
Nature, 540, 2016
|
|
5VVR
 
 | |
1Z3I
 
 | | Structure of the SWI2/SNF2 chromatin remodeling domain of eukaryotic Rad54 | | Descriptor: | SULFATE ION, ZINC ION, similar to RAD54-like | | Authors: | Thoma, N.H, Czyzewski, B.K, Alexeev, A.A, Mazin, A.V, Kowalczykowski, S.C, Pavletich, N.P. | | Deposit date: | 2005-03-12 | | Release date: | 2005-04-05 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure of the SWI2/SNF2 chromatin-remodeling domain of eukaryotic Rad54.
Nat.Struct.Mol.Biol., 12, 2005
|
|
8B3F
 
 | | Pol II-CSB-CSA-DDB1-ELOF1 | | Descriptor: | DNA damage-binding protein 1, DNA excision repair protein ERCC-6, DNA excision repair protein ERCC-8, ... | | Authors: | Kokic, G, Cramer, P. | | Deposit date: | 2022-09-16 | | Release date: | 2023-09-27 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Pol II-CSB-CSA-DDB1-ELOF1 structure.
To Be Published
|
|
8B3D
 
 | | Structure of the Pol II-TCR-ELOF1 complex. | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, DNA damage-binding protein 1, ... | | Authors: | Kokic, G, Cramer, P. | | Deposit date: | 2022-09-16 | | Release date: | 2023-09-27 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Structure of the Pol II-TCR-ELOF1 complex.
To Be Published
|
|
8C8H
 
 | |
7AOH
 
 | | Atomic structure of the poxvirus late initially transcribing complex | | Descriptor: | DNA-directed RNA polymerase, DNA-directed RNA polymerase 147 kDa polypeptide, DNA-directed RNA polymerase 18 kDa subunit, ... | | Authors: | Grimm, C, Bartuli, J, Fischer, U. | | Deposit date: | 2020-10-14 | | Release date: | 2021-10-06 | | Last modified: | 2021-10-20 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Structural basis of the complete poxvirus transcription initiation process.
Nat.Struct.Mol.Biol., 28, 2021
|
|
6BOG
 
 | | Crystal structure of RapA, a Swi2/Snf2 protein that recycles RNA polymerase during transcription | | Descriptor: | RNA polymerase-associated protein RapA, SULFATE ION | | Authors: | Shaw, G.X, Gan, J, Zhou, Y.N, Zhang, R, Joachimiak, A, Jin, D.J, Ji, X. | | Deposit date: | 2017-11-20 | | Release date: | 2017-12-13 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (3.205 Å) | | Cite: | Structure of RapA, a Swi2/Snf2 protein that recycles RNA polymerase during transcription.
Structure, 16, 2008
|
|
7EGM
 
 | | The SRM module of SWI/SNF-nucleosome complex | | Descriptor: | SWI/SNF chromatin-remodeling complex subunit SNF5, SWI/SNF chromatin-remodeling complex subunit SWI1, SWI/SNF complex subunit SWI3, ... | | Authors: | Chen, Z.C, Chen, K.J, He, Z.Y, Ye, Y.P. | | Deposit date: | 2021-03-24 | | Release date: | 2022-01-12 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structure of the SWI/SNF complex bound to the nucleosome and insights into the functional modularity.
Cell Discov, 7, 2021
|
|
6UXV
 
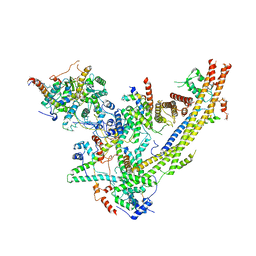 | | SWI/SNF Body Module | | Descriptor: | SWI/SNF chromatin-remodeling complex subunit SNF5, SWI/SNF chromatin-remodeling complex subunit SWI1, SWI/SNF complex subunit SWI3, ... | | Authors: | He, Y, Han, Y. | | Deposit date: | 2019-11-08 | | Release date: | 2020-03-18 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (4.7 Å) | | Cite: | Cryo-EM structure of SWI/SNF complex bound to a nucleosome.
Nature, 579, 2020
|
|
8TVY
 
 | | Cryo-EM structure of CPD lesion containing RNA Polymerase II elongation complex with Rad26 and Elf1 (closed state) | | Descriptor: | DNA (NTS), DNA (TS), DNA-directed RNA polymerase II subunit RPB1, ... | | Authors: | Sarsam, R.D, Lahiri, I, Leschziner, A.E. | | Deposit date: | 2023-08-18 | | Release date: | 2024-01-24 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Elf1 promotes Rad26's interaction with lesion-arrested Pol II for transcription-coupled repair.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
6EG3
 
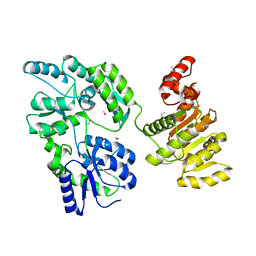 | | Crystal structure of human BRM in complex with compound 15 | | Descriptor: | 3-[(4-{[(2-chloropyridin-4-yl)carbamoyl]amino}pyridin-2-yl)ethynyl]benzoic acid, ETHANOL, Maltose/maltodextrin-binding periplasmic protein,Probable global transcription activator SNF2L2 | | Authors: | Zhu, X, Kulathila, R, Hu, T, Xie, X. | | Deposit date: | 2018-08-17 | | Release date: | 2018-10-31 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.84 Å) | | Cite: | Discovery of Orally Active Inhibitors of Brahma Homolog (BRM)/SMARCA2 ATPase Activity for the Treatment of Brahma Related Gene 1 (BRG1)/SMARCA4-Mutant Cancers.
J. Med. Chem., 61, 2018
|
|
6EG2
 
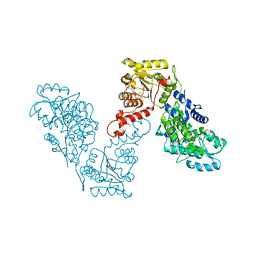 | | Crystal structure of human BRM in complex with compound 16 | | Descriptor: | ISOPROPYL ALCOHOL, Maltose/maltodextrin-binding periplasmic protein,Probable global transcription activator SNF2L2, N-(5-amino-2-chloropyridin-4-yl)-N'-(4-bromo-3-{[3-(hydroxymethyl)phenyl]ethynyl}-1,2-thiazol-5-yl)urea | | Authors: | Zhu, X, Kulathila, R, Hu, T, Xie, X. | | Deposit date: | 2018-08-17 | | Release date: | 2018-10-31 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.98 Å) | | Cite: | Discovery of Orally Active Inhibitors of Brahma Homolog (BRM)/SMARCA2 ATPase Activity for the Treatment of Brahma Related Gene 1 (BRG1)/SMARCA4-Mutant Cancers.
J. Med. Chem., 61, 2018
|
|
6G7E
 
 | |
7M8E
 
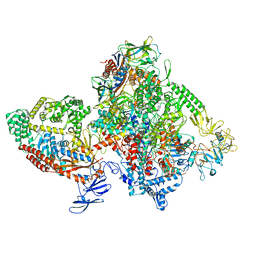 | | E.coli RNAP-RapA elongation complex | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | Shi, W, Liu, B. | | Deposit date: | 2021-03-29 | | Release date: | 2021-08-18 | | Last modified: | 2021-10-20 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural basis for activation of Swi2/Snf2 ATPase RapA by RNA polymerase.
Nucleic Acids Res., 49, 2021
|
|
7MKN
 
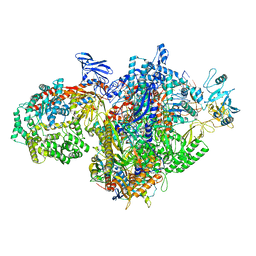 | | Escherichia coli RNA polymerase and RapA elongation complex | | Descriptor: | 5'-O-[(S)-hydroxy{[(S)-hydroxy(phosphonooxy)phosphoryl]methyl}phosphoryl]cytidine, DNA (29-MER), DNA-directed RNA polymerase subunit alpha, ... | | Authors: | Qayyum, M.Z, Murakami, K.S. | | Deposit date: | 2021-04-26 | | Release date: | 2021-06-02 | | Last modified: | 2021-12-22 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural basis of RNA polymerase recycling by the Swi2/Snf2 family of ATPase RapA in Escherichia coli.
J.Biol.Chem., 297, 2021
|
|
7MKQ
 
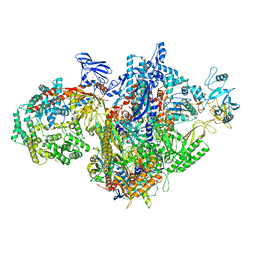 | | Escherichia coli RNA polymerase and RapA binary complex | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | Qayyum, M.Z, Murakami, K.S. | | Deposit date: | 2021-04-26 | | Release date: | 2021-06-02 | | Last modified: | 2021-12-22 | | Method: | ELECTRON MICROSCOPY (4.8 Å) | | Cite: | Structural basis of RNA polymerase recycling by the Swi2/Snf2 family of ATPase RapA in Escherichia coli.
J.Biol.Chem., 297, 2021
|
|
