8OW1
 
 | | Cryo-EM structure of the yeast Inner kinetochore bound to a CENP-A nucleosome. | | Descriptor: | C0N3, Centromere DNA-binding protein complex CBF3 subunit B, Centromere DNA-binding protein complex CBF3 subunit C, ... | | Authors: | Dendooven, T.D, Zhang, Z, Yang, J, McLaughlin, S, Schwabb, J, Scheres, S, Yatskevich, S, Barford, D. | | Deposit date: | 2023-04-26 | | Release date: | 2023-08-09 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Cryo-EM structure of the complete inner kinetochore of the budding yeast point centromere.
Sci Adv, 9, 2023
|
|
7UIO
 
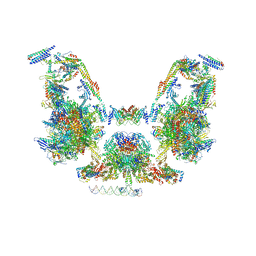 | | Mediator-PIC Early (Composite Model) | | Descriptor: | ALANINE, ASPARTIC ACID, DNA (37-MER), ... | | Authors: | Gorbea Colon, J.J, Chen, S.-F, Tsai, K.L, Murakami, K. | | Deposit date: | 2022-03-29 | | Release date: | 2023-03-01 | | Last modified: | 2023-03-08 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural basis of a transcription pre-initiation complex on a divergent promoter.
Mol.Cell, 83, 2023
|
|
7UIK
 
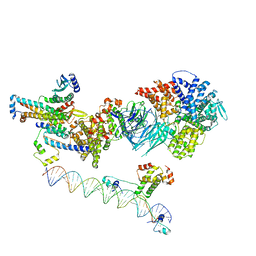 | | Mediator-PIC Early (Tail A + Upstream DNA & Activator) | | Descriptor: | DNA (37-MER), DNA (38-MER), Mediator of RNA polymerase II transcription subunit 14, ... | | Authors: | Gorbea Colon, J.J, Chen, S.-F, Tsai, K.L, Murakami, K. | | Deposit date: | 2022-03-29 | | Release date: | 2023-03-01 | | Last modified: | 2023-03-08 | | Method: | ELECTRON MICROSCOPY (7.7 Å) | | Cite: | Structural basis of a transcription pre-initiation complex on a divergent promoter.
Mol.Cell, 83, 2023
|
|
7K7G
 
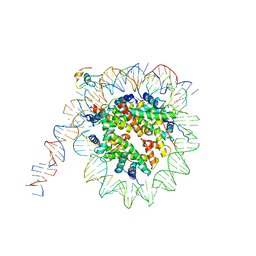 | | nucleosome and Gal4 complex | | Descriptor: | Centromere DNA-binding protein complex CBF3 subunit B, DNA (147-MER), Histone H2A.1, ... | | Authors: | Ruifang, G, Yawen, B. | | Deposit date: | 2020-09-22 | | Release date: | 2021-03-31 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Structural and dynamic mechanisms of CBF3-guided centromeric nucleosome formation.
Nat Commun, 12, 2021
|
|
7K79
 
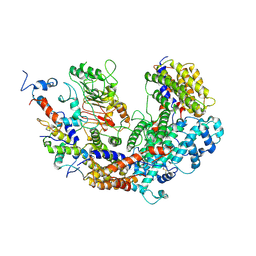 | | CBF3 | | Descriptor: | Centromere DNA-binding protein complex CBF3 subunit B, Centromere DNA-binding protein complex CBF3 subunit C, Suppressor of kinetochore protein 1 | | Authors: | Ruifang, G, Yawen, B. | | Deposit date: | 2020-09-22 | | Release date: | 2021-03-31 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structural and dynamic mechanisms of CBF3-guided centromeric nucleosome formation.
Nat Commun, 12, 2021
|
|
6V92
 
 | | RSC-NCP | | Descriptor: | Actin-like protein ARP9, Actin-related protein 7, Chromatin structure-remodeling complex protein RSC14, ... | | Authors: | Patel, A.B, Moore, C.M, Greber, B.J, Nogales, E. | | Deposit date: | 2019-12-13 | | Release date: | 2020-01-15 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (20 Å) | | Cite: | Architecture of the chromatin remodeler RSC and insights into its nucleosome engagement.
Elife, 8, 2019
|
|
6V8O
 
 | | RSC core | | Descriptor: | Chromatin structure-remodeling complex protein RSC14, Chromatin structure-remodeling complex protein RSC3, Chromatin structure-remodeling complex protein RSC30, ... | | Authors: | Patel, A.B, Moore, C.M, Greber, B.J, Nogales, E. | | Deposit date: | 2019-12-11 | | Release date: | 2020-01-15 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.07 Å) | | Cite: | Architecture of the chromatin remodeler RSC and insights into its nucleosome engagement.
Elife, 8, 2019
|
|
6P7X
 
 | | Structure of the K. lactis CBF3 core - Ndc10 D1D2 complex | | Descriptor: | Cep3, Ctf13, Ndc10, ... | | Authors: | Lee, P.D, Wei, H, Tan, D, Harrison, S.C. | | Deposit date: | 2019-06-06 | | Release date: | 2019-09-18 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Structure of the Centromere Binding Factor 3 Complex from Kluyveromyces lactis.
J.Mol.Biol., 431, 2019
|
|
6P7W
 
 | | Structure of the K. lactis CBF3 core - Ndc10 D1 complex | | Descriptor: | Cep3, Ctf13, Ndc10, ... | | Authors: | Lee, P.D, Wei, H, Tan, D, Harrison, S.C. | | Deposit date: | 2019-06-06 | | Release date: | 2019-09-18 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Structure of the Centromere Binding Factor 3 Complex from Kluyveromyces lactis.
J.Mol.Biol., 431, 2019
|
|
6P7V
 
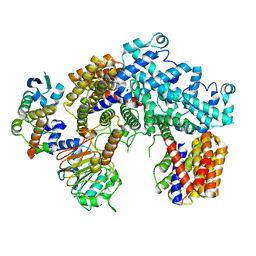 | | Structure of the K. lactis CBF3 core | | Descriptor: | Cep3, Ctf13, Skp1 | | Authors: | Lee, P.D, Wei, H, Tan, D, Harrison, S.C. | | Deposit date: | 2019-06-06 | | Release date: | 2019-09-11 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structure of the Centromere Binding Factor 3 Complex from Kluyveromyces lactis.
J.Mol.Biol., 431, 2019
|
|
6KW5
 
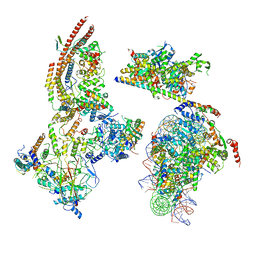 | | The ClassC RSC-Nucleosome Complex | | Descriptor: | Actin-like protein ARP9, Actin-related protein 7, Chromatin structure-remodeling complex protein RSC3, ... | | Authors: | Ye, Y.P, Wu, H, Chen, K.J, Verma, N, Cairns, B, Gao, N, Chen, Z.C. | | Deposit date: | 2019-09-06 | | Release date: | 2020-09-09 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (10.13 Å) | | Cite: | Structure of the RSC complex bound to the nucleosome
To Be Published
|
|
6KW4
 
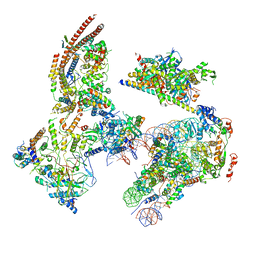 | | The ClassB RSC-Nucleosome Complex | | Descriptor: | Actin-like protein ARP9, Actin-related protein 7, Chromatin structure-remodeling complex protein RSC3, ... | | Authors: | Ye, Y.P, Wu, H, Chen, K.J, Verma, N, Cairns, B, Gao, N, Chen, Z.C. | | Deposit date: | 2019-09-06 | | Release date: | 2019-11-13 | | Last modified: | 2023-07-26 | | Method: | ELECTRON MICROSCOPY (7.55 Å) | | Cite: | Structure of the RSC complex bound to the nucleosome.
Science, 366, 2019
|
|
6KW3
 
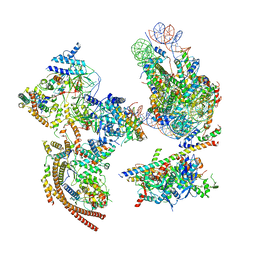 | | The ClassA RSC-Nucleosome Complex | | Descriptor: | Actin-like protein ARP9, Actin-related protein 7, Chromatin structure-remodeling complex protein RSC3, ... | | Authors: | Ye, Y.P, Wu, H, Chen, K.J, Verma, N, Cairns, B, Gao, N, Chen, Z.C. | | Deposit date: | 2019-09-05 | | Release date: | 2019-11-13 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (7.13 Å) | | Cite: | Structure of the RSC complex bound to the nucleosome.
Science, 366, 2019
|
|
6K15
 
 | | RSC substrate-recruitment module | | Descriptor: | Chromatin structure-remodeling complex protein RSC3, Chromatin structure-remodeling complex protein RSC30, Chromatin structure-remodeling complex protein RSC58, ... | | Authors: | Ye, Y.P, Wu, H, Chen, K.J, Verma, N, Cairns, B, Gao, N, Chen, Z.C. | | Deposit date: | 2019-05-09 | | Release date: | 2019-11-13 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structure of the RSC complex bound to the nucleosome.
Science, 366, 2019
|
|
6GYU
 
 | | Cryo-EM structure of the CBF3-msk complex of the budding yeast kinetochore | | Descriptor: | Centromere DNA-binding protein complex CBF3 subunit A, Centromere DNA-binding protein complex CBF3 subunit B, Centromere DNA-binding protein complex CBF3 subunit C, ... | | Authors: | Yan, K, Zhang, Z, Yang, J, McLaughlin, S.H, Barford, D. | | Deposit date: | 2018-07-02 | | Release date: | 2018-12-05 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Architecture of the CBF3-centromere complex of the budding yeast kinetochore.
Nat. Struct. Mol. Biol., 25, 2018
|
|
6GYS
 
 | | Cryo-EM structure of the CBF3-CEN3 complex of the budding yeast kinetochore | | Descriptor: | Centromere DNA-binding protein complex CBF3 subunit A, Centromere DNA-binding protein complex CBF3 subunit B, Centromere DNA-binding protein complex CBF3 subunit C, ... | | Authors: | Yan, K, Zhang, Z, Yang, J, McLaughlin, S.H, Barford, D. | | Deposit date: | 2018-07-01 | | Release date: | 2018-12-05 | | Last modified: | 2019-12-11 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Architecture of the CBF3-centromere complex of the budding yeast kinetochore.
Nat. Struct. Mol. Biol., 25, 2018
|
|
6GYP
 
 | | Cryo-EM structure of the CBF3-core-Ndc10-DBD complex of the budding yeast kinetochore | | Descriptor: | ARGININE, ASPARAGINE, Centromere DNA-binding protein complex CBF3 subunit A, ... | | Authors: | Yan, K, Zhang, Z, Yang, J, McLaughlin, S.H, Barford, D. | | Deposit date: | 2018-07-01 | | Release date: | 2018-12-05 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Architecture of the CBF3-centromere complex of the budding yeast kinetochore.
Nat. Struct. Mol. Biol., 25, 2018
|
|
6F07
 
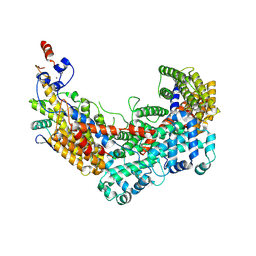 | | CBF3 Core Complex | | Descriptor: | Centromere DNA-binding protein complex CBF3 subunit B, Suppressor of kinetochore protein 1, ZINC ION | | Authors: | Leber, V, Singleton, M.R. | | Deposit date: | 2017-11-17 | | Release date: | 2017-12-13 | | Last modified: | 2019-12-11 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structural basis for assembly of the CBF3 kinetochore complex.
EMBO J., 37, 2018
|
|
3COQ
 
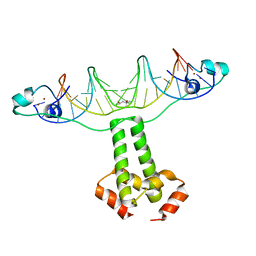 | | Structural Basis for Dimerization in DNA Recognition by Gal4 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, DNA (5'-D(*DAP*DCP*DCP*DGP*DGP*DAP*DGP*DGP*DAP*DCP*DAP*DGP*DTP*DCP*DCP*DTP*DCP*DCP*DGP*DG)-3'), DNA (5'-D(*DTP*DCP*DCP*DGP*DGP*DAP*DGP*DGP*DAP*DCP*DTP*DGP*DTP*DCP*DCP*DTP*DCP*DCP*DGP*DG)-3'), ... | | Authors: | Hong, M, Fitzgerald, M.X, Harper, S, Luo, C, Speicher, D.W. | | Deposit date: | 2008-03-29 | | Release date: | 2008-07-01 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis for dimerization in DNA recognition by gal4.
Structure, 16, 2008
|
|
3ALC
 
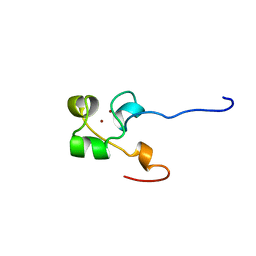 | | ETHANOL REGULON TRANSCRIPTIONAL ACTIVATOR DNA-BINDING DOMAIN FROM ASPERGILLUS NIDULANS | | Descriptor: | PROTEIN (ETHANOL REGULON TRANSCRIPTIONAL ACTIVATOR), ZINC ION | | Authors: | Cerdan, R, Cahuzac, B, Felenbok, B, Guittet, E. | | Deposit date: | 1999-03-11 | | Release date: | 2000-05-15 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | NMR solution structure of AlcR (1-60) provides insight in the unusual DNA binding properties of this zinc binuclear cluster protein.
J.Mol.Biol., 295, 2000
|
|
2HAP
 
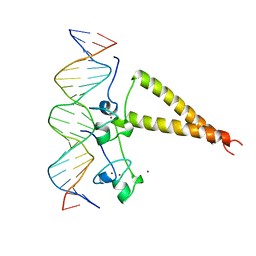 | | STRUCTURE OF A HAP1-18/DNA COMPLEX REVEALS THAT PROTEIN/DNA INTERACTIONS CAN HAVE DIRECT ALLOSTERIC EFFECTS ON TRANSCRIPTIONAL ACTIVATION | | Descriptor: | DNA (5'-D(*AP*CP*GP*CP*TP*AP*TP*TP*AP*TP*CP*GP*CP*TP*AP*TP*TP*AP*GP*T)-3'), DNA (5'-D(*AP*CP*TP*AP*AP*TP*AP*GP*CP*GP*AP*TP*AP*AP*TP*AP*GP*CP*GP*T)-3'), PROTEIN (HEME ACTIVATOR PROTEIN), ... | | Authors: | King, D.A, Zhang, L, Guarente, L, Marmorstein, R. | | Deposit date: | 1998-09-17 | | Release date: | 1999-11-10 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of HAP1-18-DNA implicates direct allosteric effect of protein-DNA interactions on transcriptional activation.
Nat.Struct.Biol., 6, 1999
|
|
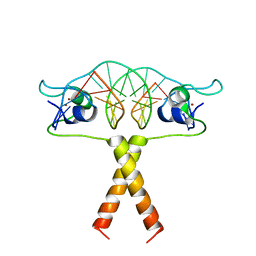
2ERG
 
 | |
2ERE
 
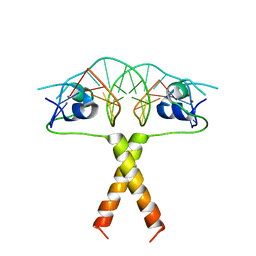 | |
2ER8
 
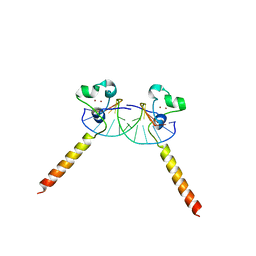 | |
2ALC
 
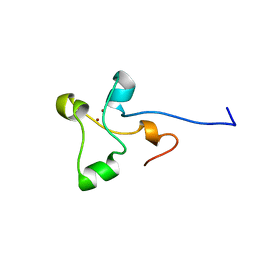 | | ETHANOL REGULON TRANSCRIPTIONAL ACTIVATOR DNA-BINDING DOMAIN FROM ASPERGILLUS NIDULANS | | Descriptor: | PROTEIN (ETHANOL REGULON TRANSCRIPTIONAL ACTIVATOR), ZINC ION | | Authors: | Cerdan, R, Cahuzac, B, Felenbok, B, Guittet, E. | | Deposit date: | 1999-01-20 | | Release date: | 2000-01-21 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | NMR solution structure of AlcR (1-60) provides insight in the unusual DNA binding properties of this zinc binuclear cluster protein.
J.Mol.Biol., 295, 2000
|
|
