8ACF
 
 | |
8ACI
 
 | |
8TYP
 
 | | Complement Protease C1s Inhibited by 6-(4-phenylpiperazin-1-yl)pyridine-3-carboximidamide | | Descriptor: | 6-(4-phenylpiperazin-1-yl)pyridine-3-carboximidamide, Complement C1s subcomponent, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Geisbrecht, B.V. | | Deposit date: | 2023-08-25 | | Release date: | 2023-11-29 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Inhibition of the C1s Protease and the Classical Complement Pathway by 6-(4-Phenylpiperazin-1-yl)Pyridine-3-Carboximidamide and Chemical Analogs.
J Immunol., 212, 2024
|
|
8SL1
 
 | | Cryo-EM structure of PAPP-A2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Judge, R.A, Stoll, V.S, Eaton, D, Hao, Q, Bratkowski, M.A. | | Deposit date: | 2023-04-20 | | Release date: | 2023-11-08 | | Method: | ELECTRON MICROSCOPY (3.13 Å) | | Cite: | Cryo-EM structure of human PAPP-A2 and mechanism of substrate recognition
Commun Chem, 6, 2023
|
|
8QK3
 
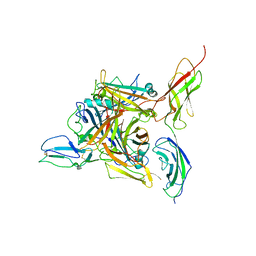 | |
8EOK
 
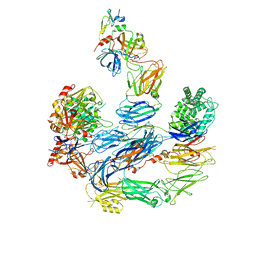 | |
8ENU
 
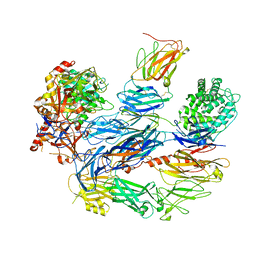 | | Structure of the C3bB proconvertase in complex with lufaxin | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Complement C3 beta chain, Complement C3b alpha' chain, ... | | Authors: | Andersen, J.F, Lei, H. | | Deposit date: | 2022-09-30 | | Release date: | 2023-08-09 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (3.22 Å) | | Cite: | A bispecific inhibitor of complement and coagulation blocks activation in complementopathy models via a novel mechanism.
Blood, 141, 2023
|
|
8CI3
 
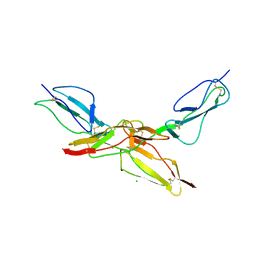 | | Structure of bovine CD46 ectodomain (SCR 1-2) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, ... | | Authors: | Aitkenhead, H, David I Stuart, D.I, El Omari, K. | | Deposit date: | 2023-02-08 | | Release date: | 2023-07-12 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Structure of Bovine CD46 Ectodomain.
Viruses, 15, 2023
|
|
8CJV
 
 | |
8GMN
 
 | | Crystal structure of human C1s in complex with inhibitor | | Descriptor: | Complement C1s subcomponent, SULFATE ION, [4-(1-aminophthalazin-6-yl)piperazin-1-yl](2-methylphenyl)methanone | | Authors: | Dougan, D.R, Lane, W. | | Deposit date: | 2023-03-26 | | Release date: | 2023-05-17 | | Last modified: | 2023-05-24 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Discovery of a Novel Series of Potent, Selective, Orally Available, and Brain-Penetrable C1s Inhibitors for Modulation of the Complement Pathway.
J.Med.Chem., 66, 2023
|
|
7ZJM
 
 | | Crystal structure of a complex between CspZ from Borrelia burgdorferi strain B408 and human FH SCR domains 6-7 | | Descriptor: | Complement factor H, CspZ, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Brangulis, K, Marcinkiewicz, A, Hart, T.M, Dupuis, A.P, Zamba Campero, M, Nowak, T.A, Stout, J.L, Akopjana, I, Kazaks, A, Bogans, J, Ciota, A.T, Kraiczy, P, Kolokotronis, S.O, Lin, Y.-P. | | Deposit date: | 2022-04-11 | | Release date: | 2023-04-19 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Structural evolution of an immune evasion determinant shapes pathogen host tropism.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8B0G
 
 | | 2C9, C5b9-CD59 structure | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CD59 glycoprotein, Complement C5, ... | | Authors: | Couves, E.C, Gardner, S, Bubeck, D. | | Deposit date: | 2022-09-07 | | Release date: | 2023-02-22 | | Last modified: | 2023-03-01 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural basis for membrane attack complex inhibition by CD59.
Nat Commun, 14, 2023
|
|
8B0F
 
 | | CryoEM structure of C5b8-CD59 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Bubeck, D, Couves, E.C, Gardner, S. | | Deposit date: | 2022-09-07 | | Release date: | 2023-02-22 | | Last modified: | 2023-03-01 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural basis for membrane attack complex inhibition by CD59.
Nat Commun, 14, 2023
|
|
8B0H
 
 | | 2C9, C5b9-CD59 cryoEM structure | | Descriptor: | CD59 glycoprotein, Complement C5, Complement component C6, ... | | Authors: | Couves, E.C, Gardner, S, Bubeck, D. | | Deposit date: | 2022-09-07 | | Release date: | 2023-02-22 | | Last modified: | 2023-03-01 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural basis for membrane attack complex inhibition by CD59.
Nat Commun, 14, 2023
|
|
8HGH
 
 | | Structure of 2:2 PAPP-A.STC2 complex | | Descriptor: | Maltose/maltodextrin-binding periplasmic protein,Pappalysin-1, Stanniocalcin-2, ZINC ION | | Authors: | Zhong, Q.H, Chu, H.L, Wang, G.P, Zhang, C, Wei, Y, Qiao, J, Hang, J. | | Deposit date: | 2022-11-14 | | Release date: | 2023-01-11 | | Method: | ELECTRON MICROSCOPY (4.16 Å) | | Cite: | Structural insights into the covalent regulation of PAPP-A activity by proMBP and STC2.
Cell Discov, 8, 2022
|
|
8HGG
 
 | | Structure of 2:2 PAPP-A.ProMBP complex | | Descriptor: | Bone marrow proteoglycan, Pappalysin-1, ZINC ION | | Authors: | Zhong, Q.H, Chu, H.L, Wang, G.P, Zhang, C, Wei, Y, Qiao, J, Hang, J. | | Deposit date: | 2022-11-14 | | Release date: | 2023-01-11 | | Method: | ELECTRON MICROSCOPY (3.64 Å) | | Cite: | Structural insights into the covalent regulation of PAPP-A activity by proMBP and STC2.
Cell Discov, 8, 2022
|
|
7Y5Q
 
 | | Structure of 1:1 PAPP-A.STC2 complex(half map) | | Descriptor: | Maltose/maltodextrin-binding periplasmic protein,Pappalysin-1, Stanniocalcin-2, ZINC ION | | Authors: | Zhong, Q.H, Chu, H.L, Wang, G.P, Zhang, C, Wei, Y, Qiao, J, Hang, J. | | Deposit date: | 2022-06-17 | | Release date: | 2023-01-11 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structural insights into the covalent regulation of PAPP-A activity by proMBP and STC2.
Cell Discov, 8, 2022
|
|
7Y5N
 
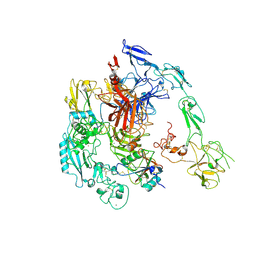 | | Structure of 1:1 PAPP-A.ProMBP complex(half map) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Bone marrow proteoglycan, ... | | Authors: | Zhong, Q.H, Chu, H.L, Wang, G.P, Zhang, C, Wei, Y, Qiao, J, Hang, J. | | Deposit date: | 2022-06-17 | | Release date: | 2023-01-11 | | Method: | ELECTRON MICROSCOPY (3.45 Å) | | Cite: | Structural insights into the covalent regulation of PAPP-A activity by proMBP and STC2.
Cell Discov, 8, 2022
|
|
7ZMZ
 
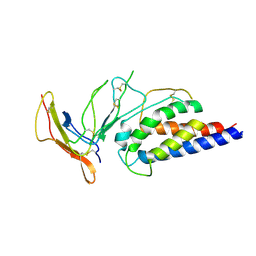 | | Engineered Interleukin 2 bound to CD25 receptor | | Descriptor: | Interleukin-2, Interleukin-2 receptor subunit alpha | | Authors: | Fyfe, P.K, Moraga, I, Gaggero, S, Mitra, S. | | Deposit date: | 2022-04-20 | | Release date: | 2022-12-14 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | IL-2 is inactivated by the acidic pH environment of tumors enabling engineering of a pH-selective mutein.
Sci Immunol, 7, 2022
|
|
8B9F
 
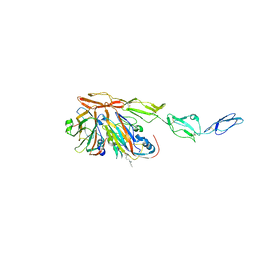 | | Structure of Echovirus 11 complexed with DAF (CD55) calculated from symmetry expansion | | Descriptor: | Complement decay-accelerating factor, Genome polyprotein, SPHINGOSINE | | Authors: | Stuart, D.I, Ren, J, Qin, L, Zhou, D. | | Deposit date: | 2022-10-05 | | Release date: | 2022-12-07 | | Last modified: | 2023-01-04 | | Method: | ELECTRON MICROSCOPY (3.93 Å) | | Cite: | Switching of Receptor Binding Poses between Closely Related Enteroviruses.
Viruses, 14, 2022
|
|
8B8R
 
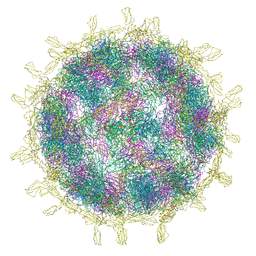 | | Complex of Echovirus 11 with its attaching receptor decay-accelerating factor (CD55) | | Descriptor: | DECAY ACCELERATING FACTOR (CD55), SPHINGOSINE, VP1, ... | | Authors: | Stuart, D.I, Ren, J, Zhou, D, Qin, L. | | Deposit date: | 2022-10-04 | | Release date: | 2022-12-07 | | Last modified: | 2023-01-04 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Switching of Receptor Binding Poses between Closely Related Enteroviruses.
Viruses, 14, 2022
|
|
7WKI
 
 | |
8A7E
 
 | | PAPP-A dimer in complex with its inhibitor STC2 | | Descriptor: | CALCIUM ION, Pappalysin-1, Stanniocalcin-2, ... | | Authors: | Kobbero, S.D, Gajhede, M, Mirza, O.A, Boesen, T, Oxvig, C. | | Deposit date: | 2022-06-20 | | Release date: | 2022-11-02 | | Method: | ELECTRON MICROSCOPY (5.02 Å) | | Cite: | Structure of the proteolytic enzyme PAPP-A with the endogenous inhibitor stanniocalcin-2 reveals its inhibitory mechanism.
Nat Commun, 13, 2022
|
|
8A7D
 
 | | Partial dimer complex of PAPP-A and its inhibitor STC2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Pappalysin-1, ... | | Authors: | Kobbero, S.D, Gajhede, M, Mirza, O.A, Boesen, T, Oxvig, C. | | Deposit date: | 2022-06-20 | | Release date: | 2022-11-02 | | Method: | ELECTRON MICROSCOPY (3.06 Å) | | Cite: | Structure of the proteolytic enzyme PAPP-A with the endogenous inhibitor stanniocalcin-2 reveals its inhibitory mechanism.
Nat Commun, 13, 2022
|
|
7UFG
 
 | | Cryo-EM structure of PAPP-A in complex with IGFBP5 | | Descriptor: | Insulin-like growth factor-binding protein 5, Pappalysin-1, ZINC ION | | Authors: | Judge, R.A, Jain, R, Hao, Q, Ouch, C, Sridar, J, Smith, C.L, Wang, J.C.K, Eaton, D. | | Deposit date: | 2022-03-22 | | Release date: | 2022-09-28 | | Method: | ELECTRON MICROSCOPY (3.28 Å) | | Cite: | Structure of the PAPP-ABP5 complex reveals mechanism of substrate recognition
Nat Commun, 13, 2022
|
|
