2UZ8
 
 | | The crystal structure of p18, human translation elongation factor 1 epsilon 1 | | Descriptor: | EUKARYOTIC TRANSLATION ELONGATION FACTOR 1 EPSILON-1, GLYCEROL | | Authors: | Kang, B.S, Kim, K.J, Kim, M.H, Oh, Y.S, Kim, S. | | Deposit date: | 2007-04-26 | | Release date: | 2008-03-25 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Determination of Three-Dimensional Structure and Residues of the Novel Tumor Suppressor Aimp3/P18 Required for the Interaction with Atm.
J.Biol.Chem., 283, 2008
|
|
5BMU
 
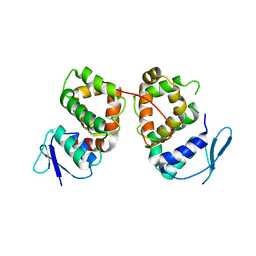 | |
4RI7
 
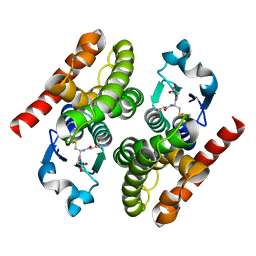 | | Crystal structure of poplar glutathione transferase F1 mutant SER 13 CYS | | Descriptor: | GLUTATHIONE, Phi class glutathione transferase GSTF1 | | Authors: | Pegeot, H, Mathiot, S, Didierjean, C, Rouhier, N. | | Deposit date: | 2014-10-05 | | Release date: | 2015-01-21 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.798 Å) | | Cite: | The poplar Phi class glutathione transferase: expression, activity and structure of GSTF1.
Front Plant Sci, 5, 2014
|
|
4RI6
 
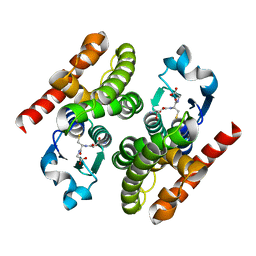 | | Crystal structure of poplar glutathione transferase F1 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, GLUTATHIONE, Phi class glutathione transferase GSTF1 | | Authors: | Pegeot, H, Koh, C.S, Didierjean, C, Rouhier, N. | | Deposit date: | 2014-10-05 | | Release date: | 2015-01-21 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.523 Å) | | Cite: | The poplar Phi class glutathione transferase: expression, activity and structure of GSTF1.
Front Plant Sci, 5, 2014
|
|
7RKA
 
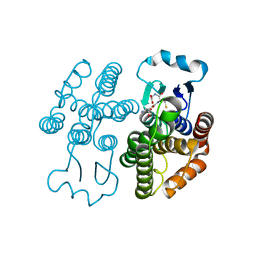 | |
7RHP
 
 | |
6V91
 
 | |
6WEG
 
 | |
5NR1
 
 | | FzlA from C. crescentus | | Descriptor: | FtsZ-binding protein FzlA | | Authors: | Szwedziak, P, Lowe, J. | | Deposit date: | 2017-04-21 | | Release date: | 2017-06-07 | | Last modified: | 2018-01-17 | | Method: | X-RAY DIFFRACTION (2.004 Å) | | Cite: | FzlA, an essential regulator of FtsZ filament curvature, controls constriction rate during Caulobacter division.
Mol. Microbiol., 107, 2018
|
|
6WMU
 
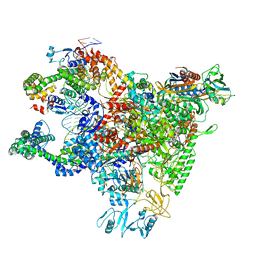 | | E. coli RNAPs70-SspA-gadA DNA complex | | Descriptor: | DNA NT-strand, DNA NT-strand downstream, DNA T-strand, ... | | Authors: | Travis, B.A, Brennan, R.G, Schumacher, M.A. | | Deposit date: | 2020-04-21 | | Release date: | 2020-11-11 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.18 Å) | | Cite: | Structural Basis for Virulence Activation of Francisella tularensis.
Mol.Cell, 81, 2021
|
|
5O00
 
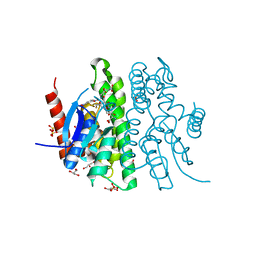 | |
5O84
 
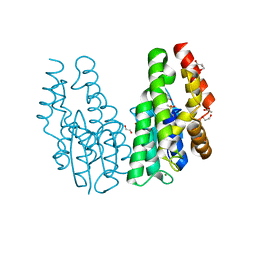 | | Glutathione S-transferase Tau 23 (partially oxidized) | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, FORMIC ACID, ... | | Authors: | Young, D.R, Van Molle, I, Tossounian, M, Messens, J. | | Deposit date: | 2017-06-12 | | Release date: | 2017-10-25 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Disulfide bond formation protects Arabidopsis thaliana glutathione transferase tau 23 from oxidative damage.
Biochim. Biophys. Acta, 1862, 2018
|
|
7OPY
 
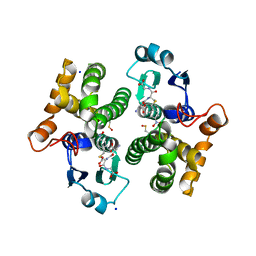 | |
7OPZ
 
 | | Camel GSTM1-1 in complex with glutathione | | Descriptor: | GLUTATHIONE, Glutathione transferase, SODIUM ION | | Authors: | Papageorgiou, A.C, Poudel, N. | | Deposit date: | 2021-06-02 | | Release date: | 2022-02-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Structural and Functional Characterization of Camelus dromedarius Glutathione Transferase M1-1.
Life, 12, 2022
|
|
7OBO
 
 | | GSTF1 from Alopecurus myosuroides | | Descriptor: | (2~{S})-2-azanyl-5-[[(2~{R})-1-(2-hydroxy-2-oxoethylamino)-3-[(7-nitro-2,1,3-benzoxadiazol-4-yl)sulfanyl]-1-oxidanylidene-propan-2-yl]amino]-5-oxidanylidene-pentanoic acid, Glutathione transferase | | Authors: | Pohl, E, Eno, R.F.M, Freitag-Pohl, S. | | Deposit date: | 2021-04-23 | | Release date: | 2022-04-20 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Flavonoid-based inhibitors of the Phi-class glutathione transferase from black-grass to combat multiple herbicide resistance.
Org.Biomol.Chem., 19, 2021
|
|
7ODM
 
 | | AmGSTF1 Y118S variant | | Descriptor: | Glutathione transferase, [(2~{S})-5-[[(2~{R})-1-(2-hydroxy-2-oxoethylamino)-1-oxidanylidene-3-sulfanyl-propan-2-yl]amino]-1-oxidanyl-1,5-bis(oxidanylidene)pentan-2-yl]azanium | | Authors: | Pohl, E, Eno, R.F.M. | | Deposit date: | 2021-04-30 | | Release date: | 2022-04-20 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Flavonoid-based inhibitors of the Phi-class glutathione transferase from black-grass to combat multiple herbicide resistance.
Org.Biomol.Chem., 19, 2021
|
|
2WJU
 
 | |
2X64
 
 | | GLUTATHIONE-S-TRANSFERASE FROM XYLELLA FASTIDIOSA | | Descriptor: | CHLORIDE ION, GLUTATHIONE, GLUTATHIONE-S-TRANSFERASE | | Authors: | Muniz, J.R.C, Rodrigues, N.C, Bleicher, L, Travensolo, R.F, Garcia, W. | | Deposit date: | 2010-02-14 | | Release date: | 2010-03-02 | | Last modified: | 2011-12-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural and Biophysical Characterization of the Recombinant Glutathione-S-Transferase from Xylella
To be Published
|
|
6YAW
 
 | | Crystal structure of human GSTA1-1 bound to the glutathione adduct of cinnamaldehyde | | Descriptor: | (2~{S})-2-azanyl-5-[[(2~{R})-1-(2-hydroxy-2-oxoethylamino)-1-oxidanylidene-3-[(1~{R})-3-oxidanylidene-1-phenyl-propyl]sulfanyl-propan-2-yl]amino]-5-oxidanylidene-pentanoic acid, GLYCEROL, Glutathione S-transferase A1 | | Authors: | Schwartz, M, Neiers, F. | | Deposit date: | 2020-03-13 | | Release date: | 2020-08-26 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Interactions Between Odorants and Glutathione Transferases in the Human Olfactory Cleft.
Chem.Senses, 45, 2020
|
|
6IY6
 
 | | Crystal structure of human cytosolic aspartyl-tRNA synthetase (DRS) in complex with glutathion-S transferase (GST) domains from Aminoacyl tRNA synthase complex-interacting multifunctional protein 2 (AIMP2) and glutamyl-prolyl-tRNA synthetase (EPRS) | | Descriptor: | Aminoacyl tRNA synthase complex-interacting multifunctional protein 2, Aspartate--tRNA ligase, cytoplasmic, ... | | Authors: | Park, S.H, Hahn, H, Han, B.W. | | Deposit date: | 2018-12-13 | | Release date: | 2019-09-11 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | The DRS-AIMP2-EPRS subcomplex acts as a pivot in the multi-tRNA synthetase complex.
Iucrj, 6, 2019
|
|
6ZJC
 
 | | Crystal structure of Equus ferus caballus glutathione transferase A3-3 in complex with glutathione and triethyltin | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, GLUTATHIONE, Glutathione S-transferase, ... | | Authors: | Skerlova, J, Ismail, A, Lindstrom, H, Sjodin, B, Mannervik, B, Stenmark, P. | | Deposit date: | 2020-06-28 | | Release date: | 2020-11-18 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural and functional analysis of the inhibition of equine glutathione transferase A3-3 by organotin endocrine disrupting pollutants.
Environ Pollut, 268, 2021
|
|
6J3F
 
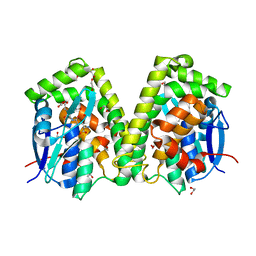 | | Crystal structure of the glutathione S-transferase, CsGST63524, of Ceriporiopsis subvermispora in complex with glutathione | | Descriptor: | 1,2-ETHANEDIOL, GLUTATHIONE, glutathione S-transferase | | Authors: | Osman, W.H.W, Mikami, B, Saka, N, Kondo, K, Nagata, T, Katahira, M. | | Deposit date: | 2019-01-04 | | Release date: | 2019-02-27 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of a serine-type glutathione S-transferase of Ceriporiopsis subvermispora and identification of the enzymatically important non-canonical residues by functional mutagenesis.
Biochem. Biophys. Res. Commun., 510, 2019
|
|
6ZJ9
 
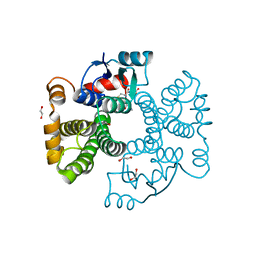 | | Crystal structure of Equus ferus caballus glutathione transferase A3-3 in complex with glutathione | | Descriptor: | 1,2-ETHANEDIOL, GLUTATHIONE, Glutathione S-transferase | | Authors: | Skerlova, J, Ismail, A, Lindstrom, H, Sjodin, B, Mannervik, B, Stenmark, P. | | Deposit date: | 2020-06-28 | | Release date: | 2020-11-18 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural and functional analysis of the inhibition of equine glutathione transferase A3-3 by organotin endocrine disrupting pollutants.
Environ Pollut, 268, 2021
|
|
6J3E
 
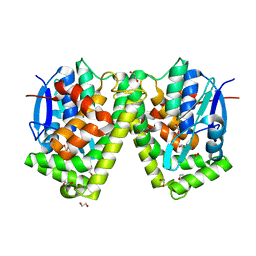 | | Crystal structure of an apo form of the glutathione S-transferase, CsGST63524, of Ceriporiopsis subvermispora | | Descriptor: | 1,2-ETHANEDIOL, glutathione S-transferase | | Authors: | Osman, W.H.W, Mikami, B, Saka, N, Kondo, K, Nagata, T, Katahira, M. | | Deposit date: | 2019-01-04 | | Release date: | 2019-02-27 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.455 Å) | | Cite: | Structure of a serine-type glutathione S-transferase of Ceriporiopsis subvermispora and identification of the enzymatically important non-canonical residues by functional mutagenesis.
Biochem. Biophys. Res. Commun., 510, 2019
|
|
6ZB6
 
 | |
