1B1G
 
 | |
1B4C
 
 | | SOLUTION STRUCTURE OF RAT APO-S100B USING DIPOLAR COUPLINGS | | Descriptor: | PROTEIN (S-100 PROTEIN, BETA CHAIN) | | Authors: | Weber, D.J, Drohat, A.C, Tjandra, N, Baldisseri, D.M. | | Deposit date: | 1998-12-17 | | Release date: | 1998-12-30 | | Last modified: | 2022-02-16 | | Method: | SOLUTION NMR | | Cite: | The use of dipolar couplings for determining the solution structure of rat apo-S100B(betabeta).
Protein Sci., 8, 1999
|
|
1BJF
 
 | |
1BOC
 
 | | THE SOLUTION STRUCTURES OF MUTANT CALBINDIN D9K'S, AS DETERMINED BY NMR, SHOW THAT THE CALCIUM BINDING SITE CAN ADOPT DIFFERENT FOLDS | | Descriptor: | CALBINDIN D9K | | Authors: | Johansson, C, Ullner, M, Drakenberg, T. | | Deposit date: | 1993-04-23 | | Release date: | 1993-10-31 | | Last modified: | 2022-02-16 | | Method: | SOLUTION NMR | | Cite: | The solution structures of mutant calbindin D9k's, as determined by NMR, show that the calcium-binding site can adopt different folds.
Biochemistry, 32, 1993
|
|
1BOD
 
 | | THE SOLUTION STRUCTURES OF MUTANT CALBINDIN D9K'S, AS DETERMINED BY NMR, SHOW THAT THE CALCIUM BINDING SITE CAN ADOPT DIFFERENT FOLDS | | Descriptor: | CALBINDIN D9K | | Authors: | Johansson, C, Ullner, M, Drakenberg, T. | | Deposit date: | 1993-04-23 | | Release date: | 1993-10-31 | | Last modified: | 2024-04-10 | | Method: | SOLUTION NMR | | Cite: | The solution structures of mutant calbindin D9k's, as determined by NMR, show that the calcium-binding site can adopt different folds.
Biochemistry, 32, 1993
|
|
1CB1
 
 | |
1CDN
 
 | |
1CFP
 
 | |
1CLB
 
 | |
1CTA
 
 | |
1CTD
 
 | |
1D1O
 
 | |
1DT7
 
 | |
1FPW
 
 | | STRUCTURE OF YEAST FREQUENIN | | Descriptor: | CALCIUM ION, CALCIUM-BINDING PROTEIN NCS-1 | | Authors: | Ames, J.B, Hendricks, K.B, Strahl, T, Huttner, I.G, Thorner, J. | | Deposit date: | 2000-08-31 | | Release date: | 2000-10-18 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Structure and calcium-binding properties of Frq1, a novel calcium sensor in the yeast Saccharomyces cerevisiae.
Biochemistry, 39, 2000
|
|
1G8I
 
 | | CRYSTAL STRUCTURE OF HUMAN FREQUENIN (NEURONAL CALCIUM SENSOR 1) | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, HEXAETHYLENE GLYCOL, ... | | Authors: | Bourne, Y, Dannenberg, J, Pollmann, V, Marchot, P, Pongs, O. | | Deposit date: | 2000-11-17 | | Release date: | 2001-05-09 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Immunocytochemical localization and crystal structure of human frequenin (neuronal calcium sensor 1).
J.Biol.Chem., 276, 2001
|
|
1GGZ
 
 | | CRYSTAL STRUCTURE OF THE CALMODULIN-LIKE PROTEIN (HCLP) FROM HUMAN EPITHELIAL CELLS | | Descriptor: | CALCIUM ION, CALMODULIN-RELATED PROTEIN NB-1 | | Authors: | Han, B.-G, Han, M, Sui, H, Yaswen, P, Walian, P.J, Jap, B.K. | | Deposit date: | 2000-10-13 | | Release date: | 2002-06-05 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of human calmodulin-like protein: insights into its functional role.
FEBS Lett., 521, 2002
|
|
1HQV
 
 | | STRUCTURE OF APOPTOSIS-LINKED PROTEIN ALG-2 | | Descriptor: | CALCIUM ION, PROGRAMMED CELL DEATH PROTEIN 6 | | Authors: | Jia, J, Tarabykina, S, Hansen, C, Berchtold, M, Cygler, M. | | Deposit date: | 2000-12-19 | | Release date: | 2001-05-02 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of apoptosis-linked protein ALG-2: insights into Ca2+-induced changes in penta-EF-hand proteins.
Structure, 9, 2001
|
|
1HT9
 
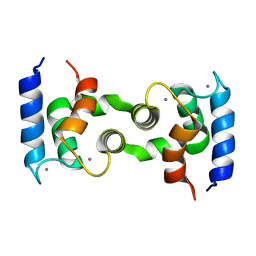 | | DOMAIN SWAPPING EF-HANDS | | Descriptor: | CALBINDIN D9K, CALCIUM ION | | Authors: | Hakansson, M, Svensson, A.L, Fast, J, Linse, S. | | Deposit date: | 2000-12-29 | | Release date: | 2001-05-09 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | An extended hydrophobic core induces EF-hand swapping.
Protein Sci., 10, 2001
|
|
1IG5
 
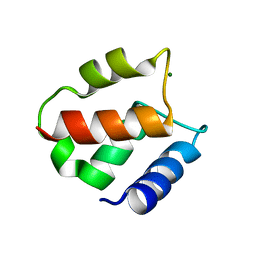 | | BOVINE CALBINDIN D9K BINDING MG2+ | | Descriptor: | MAGNESIUM ION, VITAMIN D-DEPENDENT CALCIUM-BINDING PROTEIN, INTESTINAL | | Authors: | Andersson, E.M, Svensson, L.A. | | Deposit date: | 2001-04-17 | | Release date: | 2001-04-25 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural basis for the negative allostery between Ca(2+)- and Mg(2+)-binding in the intracellular Ca(2+)-receptor calbindin D9k.
Protein Sci., 6, 1997
|
|
1IGV
 
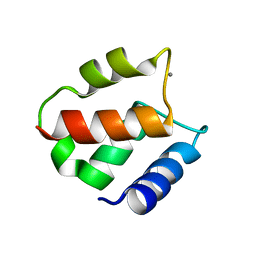 | | BOVINE CALBINDIN D9K BINDING MN2+ | | Descriptor: | MANGANESE (II) ION, VITAMIN D-DEPENDENT CALCIUM-BINDING PROTEIN, INTESTINAL | | Authors: | Andersson, E.M. | | Deposit date: | 2001-04-18 | | Release date: | 2001-04-25 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural basis for the negative allostery between Ca(2+)- and Mg(2+)-binding in the intracellular Ca(2+)-receptor calbindin D9k.
Protein Sci., 6, 1997
|
|
1IKU
 
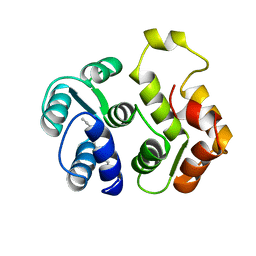 | | myristoylated recoverin in the calcium-free state, NMR, 22 structures | | Descriptor: | MYRISTIC ACID, RECOVERIN | | Authors: | Tanaka, T, Ames, J.B, Harvey, T.S, Stryer, L, Ikura, M. | | Deposit date: | 1996-01-18 | | Release date: | 1996-07-11 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Sequestration of the membrane-targeting myristoyl group of recoverin in the calcium-free state.
Nature, 376, 1995
|
|
1JBA
 
 | | UNMYRISTOYLATED GCAP-2 WITH THREE CALCIUM IONS BOUND | | Descriptor: | CALCIUM ION, PROTEIN (GUANYLATE CYCLASE ACTIVATING PROTEIN 2) | | Authors: | Ames, J.B, Dizhoor, A.M, Ikura, M, Palczewski, K, Stryer, L. | | Deposit date: | 1999-04-03 | | Release date: | 1999-12-10 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Three-dimensional structure of guanylyl cyclase activating protein-2, a calcium-sensitive modulator of photoreceptor guanylyl cyclases.
J.Biol.Chem., 274, 1999
|
|
1JC2
 
 | | COMPLEX OF THE C-DOMAIN OF TROPONIN C WITH RESIDUES 1-40 OF TROPONIN I | | Descriptor: | CALCIUM ION, TROPONIN C, SKELETAL MUSCLE | | Authors: | Mercier, P, Spyracopoulos, L, Sykes, B.D. | | Deposit date: | 2001-06-07 | | Release date: | 2001-09-14 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Structure, dynamics, and thermodynamics of the structural domain of troponin C in complex with the regulatory peptide 1-40 of troponin I
Biochemistry, 40, 2001
|
|
1JSA
 
 | | MYRISTOYLATED RECOVERIN WITH TWO CALCIUMS BOUND, NMR, 24 STRUCTURES | | Descriptor: | CALCIUM ION, MYRISTIC ACID, RECOVERIN | | Authors: | Ames, J.B, Ishima, R, Tanaka, T, Gordon, J.I, Stryer, L, Ikura, M. | | Deposit date: | 1997-06-04 | | Release date: | 1997-10-15 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Molecular mechanics of calcium-myristoyl switches.
Nature, 389, 1997
|
|
1K2H
 
 | | Three-dimensional Solution Structure of apo-S100A1. | | Descriptor: | S-100 protein, alpha chain | | Authors: | Rustandi, R.R, Baldisseri, D.M, Inman, K.G, Nizner, P, Hamilton, S.M, Landar, A, Landar, A, Zimmer, D.B, Weber, D.J. | | Deposit date: | 2001-09-27 | | Release date: | 2002-02-13 | | Last modified: | 2017-02-01 | | Method: | SOLUTION NMR | | Cite: | Three-dimensional solution structure of the calcium-signaling protein apo-S100A1 as determined by NMR.
Biochemistry, 41, 2002
|
|
