6G6K
 
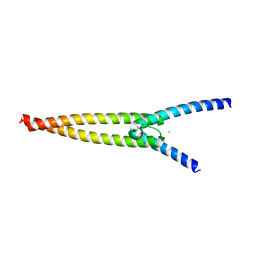 | | The crystal structures of Human MYC:MAX bHLHZip complex | | Descriptor: | CHLORIDE ION, Myc proto-oncogene protein, Protein max | | Authors: | Allen, M.D, Zinzalla, G. | | Deposit date: | 2018-04-01 | | Release date: | 2019-04-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Crystal Structures and Nuclear Magnetic Resonance Studies of the Apo Form of the c-MYC:MAX bHLHZip Complex Reveal a Helical Basic Region in the Absence of DNA.
Biochemistry, 58, 2019
|
|
6OD3
 
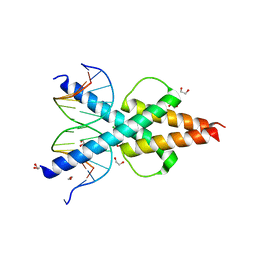 | | Human TCF4 C-terminal bHLH domain in Complex with 13-bp Oligonucleotide Containing E-box Sequence | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, DNA (5'-D(*CP*AP*TP*AP*CP*AP*CP*GP*TP*GP*TP*AP*T)-3'), ... | | Authors: | Horton, J.R, Cheng, X, Yang, J. | | Deposit date: | 2019-03-25 | | Release date: | 2019-05-29 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.494 Å) | | Cite: | Structural basis for preferential binding of human TCF4 to DNA containing 5-carboxylcytosine.
Nucleic Acids Res., 47, 2019
|
|
6U2U
 
 | |
6OD4
 
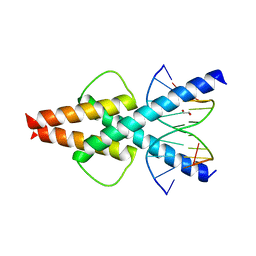 | | Human TCF4 C-terminal bHLH domain in Complex with 11-bp Oligonucleotide Containing E-box Sequence | | Descriptor: | 1,2-ETHANEDIOL, DNA (5'-D(*TP*AP*CP*AP*CP*GP*TP*GP*TP*A)-3'), Transcription factor 4 | | Authors: | Horton, J.R, Cheng, X, Yang, J. | | Deposit date: | 2019-03-25 | | Release date: | 2019-05-29 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.699 Å) | | Cite: | Structural basis for preferential binding of human TCF4 to DNA containing 5-carboxylcytosine.
Nucleic Acids Res., 47, 2019
|
|
3U5V
 
 | | Crystal structure of Max-E47 | | Descriptor: | NITRATE ION, Protein max, Transcription factor E2-alpha chimera | | Authors: | Guarne, A, Ahmadpour, F, Gloyd, M. | | Deposit date: | 2011-10-11 | | Release date: | 2012-03-21 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of the minimalist max-e47 protein chimera.
Plos One, 7, 2012
|
|
6MGM
 
 | |
1NKP
 
 | | Crystal structure of Myc-Max recognizing DNA | | Descriptor: | 5'-D(*CP*GP*AP*GP*TP*AP*GP*CP*AP*CP*GP*TP*GP*CP*TP*AP*CP*TP*C)-3', Max protein, Myc proto-oncogene protein | | Authors: | Nair, S.K, Burley, S.K. | | Deposit date: | 2003-01-03 | | Release date: | 2003-02-04 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | X-ray structures of Myc-Max and Mad-Max recognizing DNA: Molecular bases of regulation by proto-oncogenic transcription factors
Cell(Cambridge,Mass.), 112, 2003
|
|
7EOD
 
 | | MITF HLHLZ Delta AKE | | Descriptor: | GLYCEROL, Isoform M1 of Microphthalmia-associated transcription factor | | Authors: | Li, P, Liu, Z, Fang, P, Wang, J. | | Deposit date: | 2021-04-22 | | Release date: | 2022-04-27 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A unique hyperdynamic dimer interface permits small molecule perturbation of the melanoma oncoprotein MITF for melanoma therapy.
Cell Res., 33, 2023
|
|
6MGN
 
 | | mouse Id1 (51-104) - human hE47 (348-399) complex | | Descriptor: | DNA-binding protein inhibitor ID-1, Transcription factor E2-alpha | | Authors: | Benezra, R, Pavletich, N.P, Gall, A.-L, Goldgur, Y. | | Deposit date: | 2018-09-14 | | Release date: | 2019-09-18 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.901 Å) | | Cite: | A Small-Molecule Pan-Id Antagonist Inhibits Pathologic Ocular Neovascularization.
Cell Rep, 29, 2019
|
|
4ATH
 
 | | MITF apo structure | | Descriptor: | MICROPHTHALMIA-ASSOCIATED TRANSCRIPTION FACTOR, SULFATE ION | | Authors: | Pogenberg, V, Milewski, M, Wilmanns, M. | | Deposit date: | 2012-05-08 | | Release date: | 2012-12-12 | | Last modified: | 2019-07-17 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Restricted Leucine Zipper Dimerization and Specificity of DNA Recognition of the Melanocyte Master Regulator Mitf
Genes Dev., 26, 2012
|
|
5I4Z
 
 | | Structure of apo OmoMYC | | Descriptor: | CHLORIDE ION, GLYCEROL, Myc proto-oncogene protein, ... | | Authors: | Koelmel, W, Jung, L.A, Kuper, J, Eilers, M, Kisker, C. | | Deposit date: | 2016-02-13 | | Release date: | 2016-10-26 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | OmoMYC blunts promoter invasion by oncogenic MYC to inhibit gene expression characteristic of MYC-dependent tumors.
Oncogene, 36, 2017
|
|
7Y62
 
 | | Crystal structure of human TFEB HLHLZ domain | | Descriptor: | Transcription factor EB | | Authors: | Yang, G, Li, P, Lin, Y, Liu, Z, Sun, H, Zhao, Z, Fang, P, Wang, J. | | Deposit date: | 2022-06-18 | | Release date: | 2023-03-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A small-molecule drug inhibits autophagy gene expression through the central regulator TFEB.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
1NLW
 
 | | Crystal structure of Mad-Max recognizing DNA | | Descriptor: | 5'-D(*GP*AP*GP*TP*AP*GP*CP*AP*CP*GP*TP*GP*CP*TP*AP*CP*TP*C)-3', MAD PROTEIN, MAX PROTEIN | | Authors: | Nair, S.K, Burley, S.K. | | Deposit date: | 2003-01-07 | | Release date: | 2003-02-04 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | X-ray structures of Myc-Max and Mad-Max recognizing DNA: Molecular bases of regulation by proto-oncogenic transcription factors
Cell(Cambridge,Mass.), 112, 2003
|
|
6OD5
 
 | | Human TCF4 C-terminal bHLH domain in Complex with 12-bp Oligonucleotide Containing E-box Sequence with 5-carboxylcytosines | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, DNA (5'-D(*AP*(1CC)P*GP*CP*AP*CP*GP*TP*GP*(1CC)P*G)-3'), ... | | Authors: | Horton, J.R, Cheng, X, Yang, J. | | Deposit date: | 2019-03-25 | | Release date: | 2019-05-29 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural basis for preferential binding of human TCF4 to DNA containing 5-carboxylcytosine.
Nucleic Acids Res., 47, 2019
|
|
7WZ6
 
 | | Crystal structure of MyoD-E47 | | Descriptor: | Isoform E47 of Transcription factor E2-alpha, Myoblast determination protein 1 | | Authors: | Zhong, J, Huang, Y, Ma, J. | | Deposit date: | 2022-02-17 | | Release date: | 2022-06-22 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural basis of the bHLH domains of MyoD-E47 heterodimer.
Biochem.Biophys.Res.Commun., 621, 2022
|
|
6FX5
 
 | | MITF dimerization mutant | | Descriptor: | Microphthalmia-associated transcription factor, SULFATE ION | | Authors: | Pogenberg, V, Milewski, M, Wilmanns, M. | | Deposit date: | 2018-03-08 | | Release date: | 2019-03-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Mechanism of conditional partner selectivity in MITF/TFE family transcription factors with a conserved coiled coil stammer motif.
Nucleic Acids Res., 48, 2020
|
|
4AYA
 
 | | Crystal structure of ID2 HLH homodimer at 2.1A resolution | | Descriptor: | ACETATE ION, DNA-BINDING PROTEIN INHIBITOR ID-2, POTASSIUM ION | | Authors: | Wong, M.V, Jiang, S, Palasingam, P, Kolatkar, P.R. | | Deposit date: | 2012-06-19 | | Release date: | 2012-11-14 | | Method: | X-RAY DIFFRACTION (2.103 Å) | | Cite: | A Divalent Ion is Crucial in the Structure and Dominant-Negative Function of Id Proteins, a Class of Helix-Loop-Helix Transcription Regulators.
Plos One, 7, 2012
|
|
6G6L
 
 | | The crystal structures of Human MYC:MAX bHLHZip complex | | Descriptor: | Myc proto-oncogene protein, Protein max, SULFATE ION | | Authors: | Allen, M.D, Zinzalla, G. | | Deposit date: | 2018-04-01 | | Release date: | 2019-04-10 | | Last modified: | 2023-04-05 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structures and Nuclear Magnetic Resonance Studies of the Apo Form of the c-MYC:MAX bHLHZip Complex Reveal a Helical Basic Region in the Absence of DNA.
Biochemistry, 58, 2019
|
|
6G6J
 
 | | The crystal structures of Human MYC:MAX bHLHZip complex | | Descriptor: | Myc proto-oncogene protein, Protein max, SULFATE ION | | Authors: | Allen, M.D, Zinzalla, G. | | Deposit date: | 2018-04-01 | | Release date: | 2019-04-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Crystal Structures and Nuclear Magnetic Resonance Studies of the Apo Form of the c-MYC:MAX bHLHZip Complex Reveal a Helical Basic Region in the Absence of DNA.
Biochemistry, 58, 2019
|
|
4F3L
 
 | | Crystal Structure of the Heterodimeric CLOCK:BMAL1 Transcriptional Activator Complex | | Descriptor: | BMAL1b, Circadian locomoter output cycles protein kaput | | Authors: | Huang, N, Chelliah, Y, Shan, Y, Taylor, C, Yoo, S, Partch, C, Green, C.B, Zhang, H, Takahashi, J. | | Deposit date: | 2012-05-09 | | Release date: | 2012-06-06 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.268 Å) | | Cite: | Crystal structure of the heterodimeric CLOCK:BMAL1 transcriptional activator complex.
Science, 337, 2012
|
|
7D8S
 
 | | MITF bHLHLZ apo structure | | Descriptor: | Microphthalmia-associated transcription factor,Methionyl-tRNA synthetase beta subunit, SULFATE ION | | Authors: | Guo, M, Fang, P, Wang, J. | | Deposit date: | 2020-10-09 | | Release date: | 2021-10-13 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | A unique hyperdynamic dimer interface permits small molecule perturbation of the melanoma oncoprotein MITF for melanoma therapy.
Cell Res., 33, 2023
|
|
7Z5K
 
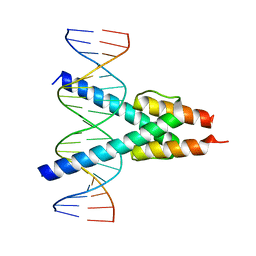 | |
1AM9
 
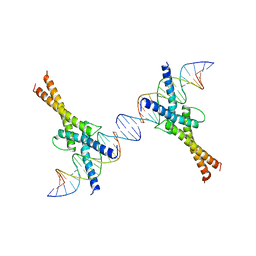 | | HUMAN SREBP-1A BOUND TO LDL RECEPTOR PROMOTER | | Descriptor: | DNA (5'-D(*CP*AP*TP*GP*AP*GP*AP*TP*CP*AP*CP*CP*CP*CP*AP*CP*T P*GP*CP*AP*A)-3'), DNA (5'-D(*TP*TP*GP*CP*AP*GP*TP*GP*GP*GP*GP*TP*GP*AP*TP*CP*T )-3'), MAGNESIUM ION, ... | | Authors: | Parraga, A, Burley, S.K. | | Deposit date: | 1997-06-25 | | Release date: | 1998-07-10 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Co-crystal structure of sterol regulatory element binding protein 1a at 2.3 A resolution.
Structure, 6, 1998
|
|
7V7L
 
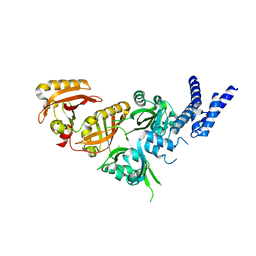 | | Crystal Structure of the Heterodimeric HIF-3a:ARNT Complex | | Descriptor: | Aryl hydrocarbon receptor nuclear translocator, Hypoxia-inducible factor 3-alpha | | Authors: | Diao, X, Ren, X, Li, F.W, Sun, X, Wu, D. | | Deposit date: | 2021-08-21 | | Release date: | 2022-05-18 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Identification of oleoylethanolamide as an endogenous ligand for HIF-3 alpha.
Nat Commun, 13, 2022
|
|
4ZP4
 
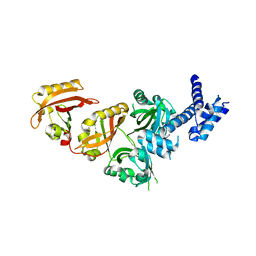 | | Crystal Structure of the Heterodimeric HIF-2a:ARNT Complex | | Descriptor: | Aryl hydrocarbon receptor nuclear translocator, Endothelial PAS domain-containing protein 1 | | Authors: | Wu, D, Potluri, N, Lu, J, Kim, Y, Rastinejad, F. | | Deposit date: | 2015-05-07 | | Release date: | 2015-08-12 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.355 Å) | | Cite: | Structural integration in hypoxia-inducible factors.
Nature, 524, 2015
|
|
