2Q9D
 
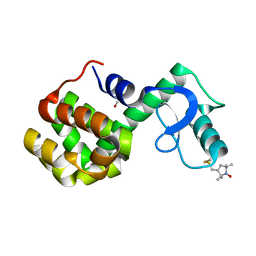 | | Structure of spin-labeled T4 lysozyme mutant A41R1 | | Descriptor: | BETA-MERCAPTOETHANOL, Lysozyme, S-[(1-oxyl-2,2,5,5-tetramethyl-2,5-dihydro-1H-pyrrol-3-yl)methyl] methanesulfonothioate | | Authors: | Guo, Z, Cascio, D, Hideg, K, Hubbell, W.L. | | Deposit date: | 2007-06-12 | | Release date: | 2007-06-26 | | Last modified: | 2023-08-30 | | Method: | EPR (1.4 Å), X-RAY DIFFRACTION | | Cite: | Structural determinants of nitroxide motion in spin-labeled proteins: Solvent-exposed sites in helix B of T4 lysozyme.
Protein Sci., 17, 2008
|
|
2Q9E
 
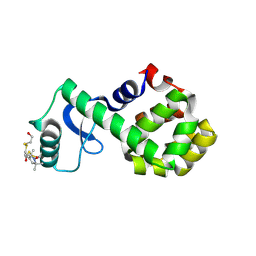 | | Structure of spin-labeled T4 lysozyme mutant S44R1 | | Descriptor: | 2-HYDROXYETHYL DISULFIDE, Lysozyme, S-[(1-oxyl-2,2,5,5-tetramethyl-2,5-dihydro-1H-pyrrol-3-yl)methyl] methanesulfonothioate | | Authors: | Guo, Z, Cascio, D, Hideg, K, Hubbell, W.L. | | Deposit date: | 2007-06-12 | | Release date: | 2007-06-26 | | Last modified: | 2023-08-30 | | Method: | EPR (2.1 Å), X-RAY DIFFRACTION | | Cite: | Structural determinants of nitroxide motion in spin-labeled proteins: Solvent-exposed sites in helix B of T4 lysozyme.
Protein Sci., 17, 2008
|
|
2RB2
 
 | |
2RBR
 
 | |
2RBS
 
 | |
2RAZ
 
 | |
2RBP
 
 | |
2RB0
 
 | | 2,6-difluorobenzylbromide complex with T4 lysozyme L99A | | Descriptor: | 2-(bromomethyl)-1,3-difluorobenzene, Lysozyme, PHOSPHATE ION | | Authors: | Graves, A.P, Boyce, S.E, Shoichet, B.K. | | Deposit date: | 2007-09-17 | | Release date: | 2008-03-18 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Rescoring docking hit lists for model cavity sites: predictions and experimental testing.
J.Mol.Biol., 377, 2008
|
|
2RBQ
 
 | |
2RBO
 
 | |
2RAY
 
 | |
2RB1
 
 | |
2RBN
 
 | |
3G3X
 
 | | Crystal structure of spin labeled T4 Lysozyme (T151R1) at 100 K | | Descriptor: | 2-HYDROXYETHYL DISULFIDE, AZIDE ION, CHLORIDE ION, ... | | Authors: | Fleissner, M.R, Cascio, D, Hubbell, W.L. | | Deposit date: | 2009-02-02 | | Release date: | 2009-05-05 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural origin of weakly ordered nitroxide motion in spin-labeled proteins.
Protein Sci., 18, 2009
|
|
3G3V
 
 | | Crystal structure of spin labeled T4 Lysozyme (V131R1) at 291 K | | Descriptor: | 2-HYDROXYETHYL DISULFIDE, AZIDE ION, CHLORIDE ION, ... | | Authors: | Fleissner, M.R, Cascio, D, Hubbell, W.L. | | Deposit date: | 2009-02-02 | | Release date: | 2009-05-05 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural origin of weakly ordered nitroxide motion in spin-labeled proteins.
Protein Sci., 18, 2009
|
|
3G3W
 
 | | Crystal structure of spin labeled T4 Lysozyme (T151R1) at 291 K | | Descriptor: | 2-HYDROXYETHYL DISULFIDE, AZIDE ION, CHLORIDE ION, ... | | Authors: | Fleissner, M.R, Cascio, D, Hubbell, W.L. | | Deposit date: | 2009-02-02 | | Release date: | 2009-05-05 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural origin of weakly ordered nitroxide motion in spin-labeled proteins.
Protein Sci., 18, 2009
|
|
3GUP
 
 | |
3GUN
 
 | |
3GUJ
 
 | |
3GUO
 
 | |
3GUM
 
 | |
3GUI
 
 | |
3GUL
 
 | |
3GUK
 
 | |
3C8R
 
 | |
