2PWG
 
 | | Crystal Structure of the Trehalulose Synthase MutB From Pseudomonas Mesoacidophila MX-45 Complexed to the Inhibitor Castanospermine | | Descriptor: | CALCIUM ION, CASTANOSPERMINE, Sucrose isomerase | | Authors: | Ravaud, S, Robert, X, Haser, R, Aghajari, N. | | Deposit date: | 2007-05-11 | | Release date: | 2007-06-26 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Trehalulose synthase native and carbohydrate complexed structures provide insights into sucrose isomerization.
J.Biol.Chem., 61, 2007
|
|
2PWH
 
 | | Crystal structure of the trehalulose synthase MutB from Pseudomonas mesoacidophila MX-45 | | Descriptor: | CALCIUM ION, Sucrose isomerase | | Authors: | Ravaud, S, Robert, X, Haser, R, Aghajari, N. | | Deposit date: | 2007-05-11 | | Release date: | 2007-06-26 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Trehalulose synthase native and carbohydrate complexed structures provide insights into sucrose isomerization.
J.Biol.Chem., 61, 2007
|
|
2QPS
 
 | | "Sugar tongs" mutant Y380A in complex with acarbose | | Descriptor: | Alpha-amylase type A isozyme, CALCIUM ION | | Authors: | Aghajari, N, Jensen, M.H, Tranier, S, Haser, R. | | Deposit date: | 2007-07-25 | | Release date: | 2008-02-12 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The 'pair of sugar tongs' site on the non-catalytic domain C of barley alpha-amylase participates in substrate binding and activity
Febs J., 274, 2007
|
|
2QMK
 
 | | Human pancreatic alpha-amylase complexed with nitrite | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, NITRITE ION, ... | | Authors: | Williams, L.K, Maurus, R, Brayer, G.D. | | Deposit date: | 2007-07-16 | | Release date: | 2008-03-25 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Alternative catalytic anions differentially modulate human alpha-amylase activity and specificity
Biochemistry, 47, 2008
|
|
2QV4
 
 | | Human pancreatic alpha-amylase complexed with nitrite and acarbose | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 4,6-dideoxy-4-{[(1S,4R,5R,6S)-4-{[alpha-D-glucopyranosyl-(1->4)-alpha-D-glucopyranosyl-(1->4)-alpha-D-glucopyranosyl]oxy}-5,6-dihydroxy-3-(hydroxymethyl)cyclohex-2-en-1-yl]amino}-alpha-D-glucopyranose, CALCIUM ION, ... | | Authors: | Williams, L.K, Maurus, R, Brayer, G.D. | | Deposit date: | 2007-08-07 | | Release date: | 2008-03-25 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Alternative catalytic anions differentially modulate human alpha-amylase activity and specificity
Biochemistry, 47, 2008
|
|
2QPU
 
 | | Sugar tongs mutant S378P in complex with acarbose | | Descriptor: | 1,2-ETHANEDIOL, 1,5-anhydro-4-O-(4,6-dideoxy-4-{[(1S,2S,3S,4R,5S,6R)-2,3,4,6-tetrahydroxy-5-methylcyclohexyl]amino}-alpha-D-glucopyranosyl)-D-glucitol, 4,6-dideoxy-4-{[(1S,4R,5S,6S)-4,5,6-trihydroxy-3-(hydroxymethyl)cyclohex-2-en-1-yl]amino}-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-beta-D-glucopyranose, ... | | Authors: | Aghajari, N, Jensen, M.H, Tranier, S, Haser, R. | | Deposit date: | 2007-07-25 | | Release date: | 2008-07-01 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The 'pair of sugar tongs' site on the non-catalytic domain C of barley alpha-amylase participates in substrate binding and activity
Febs J., 274, 2007
|
|
2TAA
 
 | | STRUCTURE AND POSSIBLE CATALYTIC RESIDUES OF TAKA-AMYLASE A | | Descriptor: | CALCIUM ION, TAKA-AMYLASE A | | Authors: | Kusunoki, M, Matsuura, Y, Tanaka, N, Kakudo, M. | | Deposit date: | 1982-10-18 | | Release date: | 1982-10-21 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure and possible catalytic residues of Taka-amylase A
J.Biochem.(Tokyo), 95, 1984
|
|
1B2Y
 
 | |
1AQM
 
 | | ALPHA-AMYLASE FROM ALTEROMONAS HALOPLANCTIS COMPLEXED WITH TRIS | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ALPHA-AMYLASE, CALCIUM ION, ... | | Authors: | Aghajari, N, Haser, R. | | Deposit date: | 1997-07-31 | | Release date: | 1999-03-02 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structures of the psychrophilic alpha-amylase from Alteromonas haloplanctis in its native form and complexed with an inhibitor.
Protein Sci., 7, 1998
|
|
1BAG
 
 | | ALPHA-AMYLASE FROM BACILLUS SUBTILIS COMPLEXED WITH MALTOPENTAOSE | | Descriptor: | ALPHA-1,4-GLUCAN-4-GLUCANOHYDROLASE, CALCIUM ION, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Fujimoto, Z, Mizuno, H, Takase, K, Doui, N. | | Deposit date: | 1998-01-30 | | Release date: | 1998-10-21 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of a catalytic-site mutant alpha-amylase from Bacillus subtilis complexed with maltopentaose.
J.Mol.Biol., 277, 1998
|
|
1AQH
 
 | | ALPHA-AMYLASE FROM ALTEROMONAS HALOPLANCTIS | | Descriptor: | ALPHA-AMYLASE, CALCIUM ION, CHLORIDE ION | | Authors: | Aghajari, N, Haser, R. | | Deposit date: | 1997-07-30 | | Release date: | 1999-02-16 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structures of the psychrophilic alpha-amylase from Alteromonas haloplanctis in its native form and complexed with an inhibitor.
Protein Sci., 7, 1998
|
|
7LV6
 
 | | The structure of MalL mutant enzyme S536R from Bacillus subtilis | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CALCIUM ION, GLYCEROL, ... | | Authors: | Hamill, C.J, Prentice, E.J, Bahl, C.D, Truebridge, I.S, Arcus, V.L. | | Deposit date: | 2021-02-24 | | Release date: | 2022-03-09 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Urea binding to guide rational design of mutations that influence enzyme dynamics
To Be Published
|
|
1BF2
 
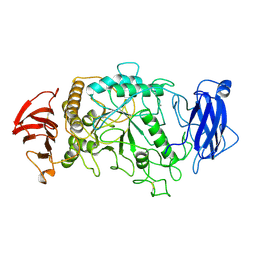 | | STRUCTURE OF PSEUDOMONAS ISOAMYLASE | | Descriptor: | CALCIUM ION, ISOAMYLASE | | Authors: | Katsuya, Y, Mezaki, Y, Kubota, M, Matsuura, Y. | | Deposit date: | 1998-05-26 | | Release date: | 1998-08-12 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Three-dimensional structure of Pseudomonas isoamylase at 2.2 A resolution.
J.Mol.Biol., 281, 1998
|
|
1BG9
 
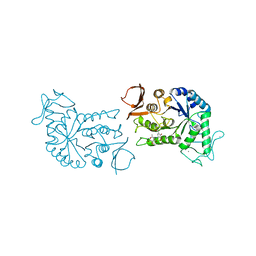 | | BARLEY ALPHA-AMYLASE WITH SUBSTRATE ANALOGUE ACARBOSE | | Descriptor: | 1,4-ALPHA-D-GLUCAN GLUCANOHYDROLASE, 4,6-dideoxy-4-{[(1S,4S,5S,6S)-4,5,6-trihydroxy-3-(hydroxymethyl)cyclohex-2-en-1-yl]amino}-beta-D-glucopyranose, 4,6-dideoxy-4-{[(1S,5R,6S)-3-formyl-5,6-dihydroxy-4-oxocyclohex-2-en-1-yl]amino}-alpha-D-xylo-hex-5-enopyranose-(1-4)-beta-D-glucopyranose, ... | | Authors: | Kadziola, A, Haser, R. | | Deposit date: | 1998-06-05 | | Release date: | 1999-06-15 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Molecular structure of a barley alpha-amylase-inhibitor complex: implications for starch binding and catalysis.
J.Mol.Biol., 278, 1998
|
|
7ML5
 
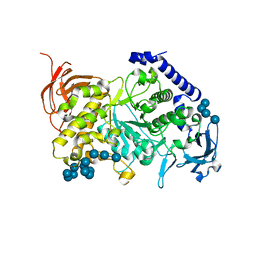 | | Structure of the Starch Branching Enzyme I (BEI) complexed with maltododecaose from Oryza sativa L | | Descriptor: | Isoform 2 of 1,4-alpha-glucan-branching enzyme, chloroplastic/amyloplastic, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, ... | | Authors: | Nayebi Gavgani, H, Fawaz, R, Geiger, J.H. | | Deposit date: | 2021-04-27 | | Release date: | 2021-11-17 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | A structural explanation for the mechanism and specificity of plant branching enzymes I and IIb.
J.Biol.Chem., 298, 2021
|
|
1BVN
 
 | | PIG PANCREATIC ALPHA-AMYLASE IN COMPLEX WITH THE PROTEINACEOUS INHIBITOR TENDAMISTAT | | Descriptor: | CALCIUM ION, CHLORIDE ION, PROTEIN (ALPHA-AMYLASE), ... | | Authors: | Machius, M, Wiegand, G, Epp, O, Huber, R. | | Deposit date: | 1998-09-16 | | Release date: | 1998-09-23 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The crystal structure of porcine pancreatic alpha-amylase in complex with the microbial inhibitor Tendamistat.
J.Mol.Biol., 247, 1995
|
|
1BVZ
 
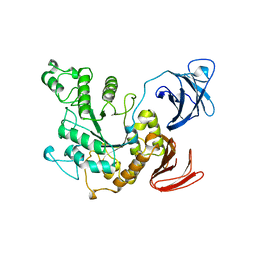 | | ALPHA-AMYLASE II (TVAII) FROM THERMOACTINOMYCES VULGARIS R-47 | | Descriptor: | PROTEIN (ALPHA-AMYLASE II) | | Authors: | Kamitori, S, Kondo, S, Okuyama, K, Yokota, T, Shimura, Y, Tonozuka, T, Sakano, Y. | | Deposit date: | 1998-09-22 | | Release date: | 1999-03-02 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of Thermoactinomyces vulgaris R-47 alpha-amylase II (TVAII) hydrolyzing cyclodextrins and pullulan at 2.6 A resolution.
J.Mol.Biol., 287, 1999
|
|
1BSI
 
 | | HUMAN PANCREATIC ALPHA-AMYLASE FROM PICHIA PASTORIS, GLYCOSYLATED PROTEIN | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ALPHA-AMYLASE, CALCIUM ION, ... | | Authors: | Rydberg, E.H, Sidhu, G, Vo, H.C, Hewitt, J, Cote, H.C.F, Wang, Y, Numao, S, Macgillivray, R.T.A, Overall, C.M, Brayer, G.D, Withers, S.G. | | Deposit date: | 1998-08-28 | | Release date: | 1999-05-18 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Cloning, mutagenesis, and structural analysis of human pancreatic alpha-amylase expressed in Pichia pastoris.
Protein Sci., 8, 1999
|
|
1C8Q
 
 | |
1CDG
 
 | | NUCLEOTIDE SEQUENCE AND X-RAY STRUCTURE OF CYCLODEXTRIN GLYCOSYLTRANSFERASE FROM BACILLUS CIRCULANS STRAIN 251 IN A MALTOSE-DEPENDENT CRYSTAL FORM | | Descriptor: | CALCIUM ION, CYCLODEXTRIN GLYCOSYL-TRANSFERASE, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Lawson, C.L, Van Montfort, R, Strokopytov, B.V, Kalk, K.H, Rozeboom, H.J, Dijkstra, B.W. | | Deposit date: | 1993-08-02 | | Release date: | 1994-01-31 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Nucleotide sequence and X-ray structure of cyclodextrin glycosyltransferase from Bacillus circulans strain 251 in a maltose-dependent crystal form.
J.Mol.Biol., 236, 1994
|
|
7NF8
 
 | | Ovine (b0,+AT-rBAT)2 hetero-tetramer, asymmetric unit, rigid-body fitted | | Descriptor: | 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Lee, Y, Kuehlbrandt, W. | | Deposit date: | 2021-02-05 | | Release date: | 2022-01-19 | | Last modified: | 2022-06-08 | | Method: | ELECTRON MICROSCOPY (2.83 Å) | | Cite: | Ca 2+ -mediated higher-order assembly of heterodimers in amino acid transport system b 0,+ biogenesis and cystinuria.
Nat Commun, 13, 2022
|
|
7NF6
 
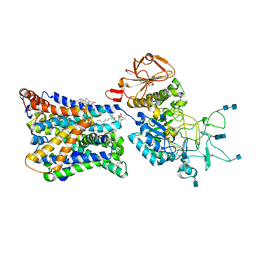 | | Ovine b0,+AT-rBAT heterodimer | | Descriptor: | 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Lee, Y, Kuehlbrandt, W. | | Deposit date: | 2021-02-05 | | Release date: | 2022-01-19 | | Last modified: | 2022-06-01 | | Method: | ELECTRON MICROSCOPY (3.05 Å) | | Cite: | Ca 2+ -mediated higher-order assembly of heterodimers in amino acid transport system b 0,+ biogenesis and cystinuria.
Nat Commun, 13, 2022
|
|
7P4W
 
 | |
7P9U
 
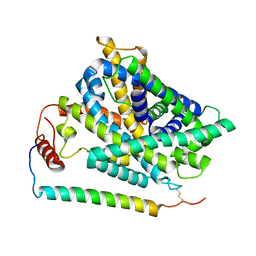 | | Cryo EM structure of System XC- in complex with glutamate | | Descriptor: | 4F2 cell-surface antigen heavy chain, Cystine/glutamate transporter, GLUTAMIC ACID | | Authors: | Parker, J.L, Deme, J.C, Lea, S.M, Newstead, S. | | Deposit date: | 2021-07-28 | | Release date: | 2021-11-17 | | Last modified: | 2022-02-02 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Molecular basis for redox control by the human cystine/glutamate antiporter system xc .
Nat Commun, 12, 2021
|
|
7P9V
 
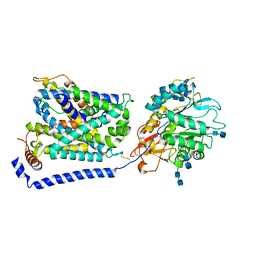 | | Cryo EM structure of System XC- | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 4F2 cell-surface antigen heavy chain, Cystine/glutamate transporter | | Authors: | Parker, J.L, Deme, J.C, Lea, S.M, Newstead, S. | | Deposit date: | 2021-07-28 | | Release date: | 2021-11-17 | | Last modified: | 2022-02-02 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Molecular basis for redox control by the human cystine/glutamate antiporter system xc .
Nat Commun, 12, 2021
|
|
