1KCK
 
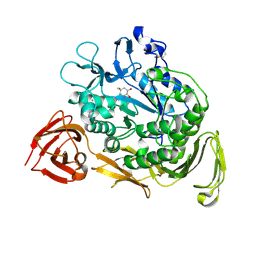 | | Bacillus circulans strain 251 Cyclodextrin glycosyl transferase mutant N193G | | Descriptor: | 1-AMINO-2,3-DIHYDROXY-5-HYDROXYMETHYL CYCLOHEX-5-ENE, CALCIUM ION, CYCLODEXTRIN GLYCOSYLTRANSFERASE, ... | | Authors: | Rozeboom, H.J, Uitdehaag, J.C.M, Dijkstra, B.W. | | Deposit date: | 2001-11-09 | | Release date: | 2002-01-16 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | The remote substrate binding subsite -6 in cyclodextrin-glycosyltransferase controls the transferase activity of the enzyme via an induced-fit mechanism.
J.Biol.Chem., 277, 2002
|
|
1KXH
 
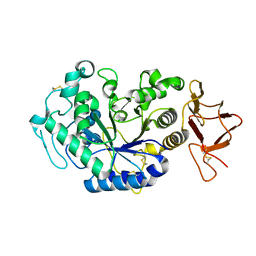 | | Crystal structure of the complex between an inactive mutant of psychrophilic alpha-amylase (D174N) and acarbose | | Descriptor: | 4,6-dideoxy-4-{[(1S,4R,5S,6S)-4,5,6-trihydroxy-3-(hydroxymethyl)cyclohex-2-en-1-yl]amino}-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Aghajari, N, Haser, R. | | Deposit date: | 2002-01-31 | | Release date: | 2002-06-19 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystallographic evidence of a transglycosylation reaction: ternary complexes of a psychrophilic alpha-amylase.
Biochemistry, 41
|
|
1KCL
 
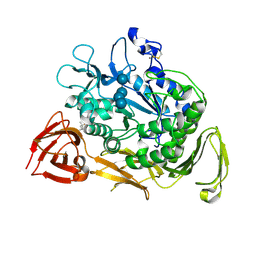 | | Bacillus ciruclans strain 251 Cyclodextrin glycosyl transferase mutant G179L | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CALCIUM ION, Cyclodextrin glycosyltransferase, ... | | Authors: | Rozeboom, H.J, Uitdehaag, J.C.M, Dijkstra, B.W. | | Deposit date: | 2001-11-09 | | Release date: | 2002-01-16 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | The remote substrate binding subsite -6 in cyclodextrin-glycosyltransferase controls the transferase activity of the enzyme via an induced-fit mechanism.
J.Biol.Chem., 277, 2002
|
|
1KXT
 
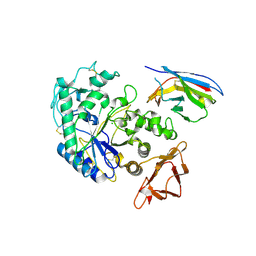 | | Camelid VHH Domains in Complex with Porcine Pancreatic alpha-Amylase | | Descriptor: | ALPHA-AMYLASE, PANCREATIC, CALCIUM ION, ... | | Authors: | Desmyter, A, Spinelli, S, Payan, F, Lauwereys, M, Wyns, L, Muyldermans, S, Cambillau, C. | | Deposit date: | 2002-02-01 | | Release date: | 2002-06-19 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Three camelid VHH domains in complex with porcine pancreatic alpha-amylase. Inhibition and versatility of binding topology.
J.Biol.Chem., 277, 2002
|
|
1KGW
 
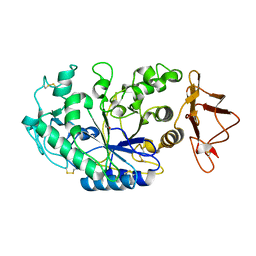 | | THREE DIMENSIONAL STRUCTURE ANALYSIS OF THE R337Q VARIANT OF HUMAN PANCREATIC ALPHA-MYLASE | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ALPHA-AMYLASE, PANCREATIC, ... | | Authors: | Numao, S, Maurus, R, Sidhu, G, Wang, Y, Overall, C.M, Brayer, G.D, Withers, S.G. | | Deposit date: | 2001-11-28 | | Release date: | 2002-01-16 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Probing the role of the chloride ion in the mechanism of human pancreatic alpha-amylase.
Biochemistry, 41, 2002
|
|
1KGX
 
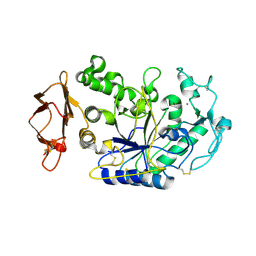 | | Three Dimensional Structure Analysis of the R195Q Variant of Human Pancreatic Alpha Amylase | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ALPHA-AMYLASE, PANCREATIC, ... | | Authors: | Numao, S, Maurus, R, Sidhu, G, Wang, Y, Overall, C.M, Brayer, G.D, Withers, S.G. | | Deposit date: | 2001-11-28 | | Release date: | 2002-01-16 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Probing the role of the chloride ion in the mechanism of human pancreatic alpha-amylase.
Biochemistry, 41, 2002
|
|
1KGU
 
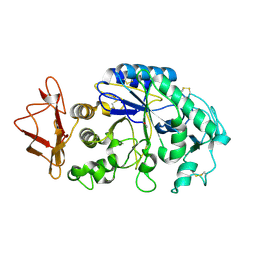 | | THREE DIMENSIONAL STRUCTURE ANALYSIS OF THE R337A VARIANT OF HUMAN PANCREATIC ALPHA-AMYLASE | | Descriptor: | ALPHA-AMYLASE, PANCREATIC, CALCIUM ION | | Authors: | Numao, S, Maurus, R, Sidhu, G, Wang, Y, Overall, C.M, Brayer, G.D, Withers, S.G. | | Deposit date: | 2001-11-28 | | Release date: | 2002-01-16 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Probing the role of the chloride ion in the mechanism of human pancreatic alpha-amylase.
Biochemistry, 41, 2002
|
|
1KXQ
 
 | | Camelid VHH Domain in Complex with Porcine Pancreatic alpha-Amylase | | Descriptor: | CALCIUM ION, CHLORIDE ION, alpha-amylase, ... | | Authors: | Desmyter, A, Spinelli, S, Payan, F, Lauwereys, M, Wyns, L, Muyldermans, S, Cambillau, C. | | Deposit date: | 2002-02-01 | | Release date: | 2002-06-19 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Three camelid VHH domains in complex with porcine pancreatic alpha-amylase. Inhibition and versatility of binding topology.
J.Biol.Chem., 277, 2002
|
|
1KXV
 
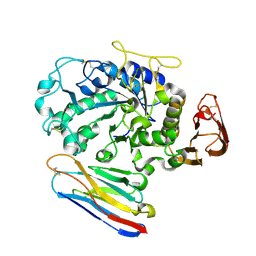 | | Camelid VHH Domains in Complex with Porcine Pancreatic alpha-Amylase | | Descriptor: | ALPHA-AMYLASE, PANCREATIC, CAMELID VHH DOMAIN CAB10 | | Authors: | Desmyter, A, Spinelli, S, Payan, F, Lauwereys, M, Wyns, L, Muyldermans, S, Cambillau, C. | | Deposit date: | 2002-02-01 | | Release date: | 2002-06-19 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Three camelid VHH domains in complex with porcine pancreatic alpha-amylase. Inhibition and versatility of binding topology.
J.Biol.Chem., 277, 2002
|
|
1SMD
 
 | | HUMAN SALIVARY AMYLASE | | Descriptor: | AMYLASE, CALCIUM ION, CHLORIDE ION | | Authors: | Ramasubbu, N. | | Deposit date: | 1996-01-24 | | Release date: | 1996-07-11 | | Last modified: | 2019-12-25 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure of human salivary alpha-amylase at 1.6 A resolution: implications for its role in the oral cavity.
Acta Crystallogr.,Sect.D, 52, 1996
|
|
1TCM
 
 | |
1TMQ
 
 | | STRUCTURE OF TENEBRIO MOLITOR LARVAL ALPHA-AMYLASE IN COMPLEX WITH RAGI BIFUNCTIONAL INHIBITOR | | Descriptor: | CALCIUM ION, CHLORIDE ION, PROTEIN (ALPHA-AMYLASE), ... | | Authors: | Gomis-Rueth, F.X, Strobl, S, Glockshuber, R. | | Deposit date: | 1998-01-13 | | Release date: | 1999-03-02 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A novel strategy for inhibition of alpha-amylases: yellow meal worm alpha-amylase in complex with the Ragi bifunctional inhibitor at 2.5 A resolution.
Structure, 6, 1998
|
|
1U2Y
 
 | | In situ extension as an approach for identifying novel alpha-amylase inhibitors, structure containing D-gluconhydroximo-1,5-lactam | | Descriptor: | (2S,3S,4R,5R)-6-(HYDROXYAMINO)-2-(HYDROXYMETHYL)-2,3,4,5-TETRAHYDROPYRIDINE-3,4,5-TRIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, Alpha-amylase, ... | | Authors: | Numao, S, Li, C, Damager, I, Wrodnigg, T.M, Begum, A, Overall, C.M, Brayer, G.D, Withers, S.G. | | Deposit date: | 2004-07-20 | | Release date: | 2004-09-07 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | In Situ Extension as an Approach for Identifying Novel alpha-Amylase Inhibitors.
J.Biol.Chem., 279, 2004
|
|
1UA7
 
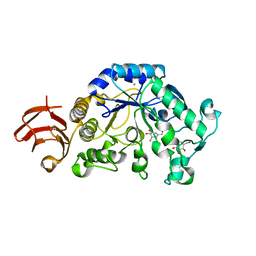 | | Crystal Structure Analysis of Alpha-Amylase from Bacillus Subtilis complexed with Acarbose | | Descriptor: | 4,6-dideoxy-alpha-D-xylo-hexopyranose-(1-4)-alpha-D-glucopyranose, 6-AMINO-4-HYDROXYMETHYL-CYCLOHEX-4-ENE-1,2,3-TRIOL, Alpha-amylase, ... | | Authors: | Kagawa, M, Fujimoto, Z, Momma, M, Takase, K, Mizuno, H. | | Deposit date: | 2003-03-03 | | Release date: | 2004-05-18 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Crystal structure of Bacillus subtilis alpha-amylase in complex with acarbose
J.BACTERIOL., 185, 2003
|
|
1UA3
 
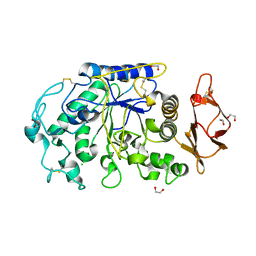 | |
1U30
 
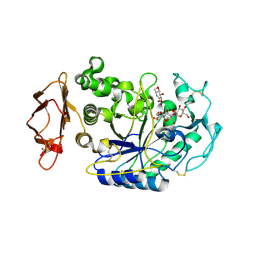 | | In situ extension as an approach for identifying novel alpha-amylase inhibitors, structure containing maltosyl-alpha (1,4)-D-gluconhydroximo-1,5-lactam | | Descriptor: | (2S,3S,4R,5R)-6-(HYDROXYAMINO)-2-(HYDROXYMETHYL)-2,3,4,5-TETRAHYDROPYRIDINE-3,4,5-TRIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, Alpha-amylase, ... | | Authors: | Numao, S, Li, C, Damager, I, Wrodnigg, T.M, Begum, A, Overall, C.M, Brayer, G.D, Withers, S.G. | | Deposit date: | 2004-07-20 | | Release date: | 2004-09-07 | | Last modified: | 2020-11-11 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | In Situ Extension as an Approach for Identifying Novel alpha-Amylase Inhibitors.
J.Biol.Chem., 279, 2004
|
|
1U33
 
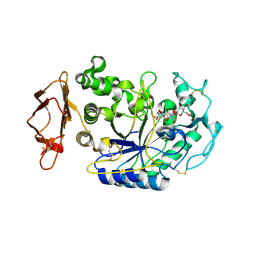 | | In situ extension as an approach for identifying novel alpha-amylase inhibitors | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 4'-O-METHYL-MALTOSYL-ALPHA (1,4)-(Z, 3S,4S,5R,6R)-3,4,5-TRIHYDROXY-6-HYDROXYMETHYL-PIPERIDIN-2-ONE, ... | | Authors: | Numao, S, Li, C, Damager, I, Wrodnigg, T.M, Begum, A, Overall, C.M, Brayer, G.D, Withers, S.G. | | Deposit date: | 2004-07-20 | | Release date: | 2004-09-07 | | Last modified: | 2020-11-11 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | In Situ Extension as an Approach for Identifying Novel alpha-Amylase Inhibitors.
J.Biol.Chem., 279, 2004
|
|
1UKQ
 
 | | Crystal structure of cyclodextrin glucanotransferase complexed with a pseudo-maltotetraose derived from acarbose | | Descriptor: | 6-AMINO-4-HYDROXYMETHYL-CYCLOHEX-4-ENE-1,2,3-TRIOL, CALCIUM ION, Cyclomaltodextrin glucanotransferase, ... | | Authors: | Haga, K, Kanai, R, Sakamoto, O, Harata, K, Yamane, K. | | Deposit date: | 2003-09-01 | | Release date: | 2004-02-24 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Effects of Essential Carbohydrate/Aromatic Stacking Interaction with Tyr100 and Phe259 on Substrate Binding of Cyclodextrin Glycosyltransferase from Alkalophilic Bacillus sp. 1011
J.Biochem.(Tokyo), 134, 2003
|
|
1UKS
 
 | | Crystal structure of F183L/F259L mutant cyclodextrin glucanotransferase complexed with a pseudo-maltotetraose derived from acarbose | | Descriptor: | 4,6-dideoxy-alpha-D-xylo-hexopyranose-(1-4)-beta-D-galactopyranose, 6-AMINO-4-HYDROXYMETHYL-CYCLOHEX-4-ENE-1,2,3-TRIOL, CALCIUM ION, ... | | Authors: | Haga, K, Kanai, R, Sakamoto, O, Harata, K, Yamane, K. | | Deposit date: | 2003-09-01 | | Release date: | 2004-02-24 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Effects of Essential Carbohydrate/Aromatic Stacking Interaction with Tyr100 and Phe259 on Substrate Binding of Cyclodextrin Glycosyltransferase from Alkalophilic Bacillus sp. 1011
J.Biochem.(Tokyo), 134, 2003
|
|
1UKT
 
 | | Crystal structure of Y100L mutant cyclodextrin glucanotransferase compexed with an acarbose | | Descriptor: | 4,6-dideoxy-alpha-D-xylo-hexopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-beta-D-galactopyranose, 6-AMINO-4-HYDROXYMETHYL-CYCLOHEX-4-ENE-1,2,3-TRIOL, CALCIUM ION, ... | | Authors: | Haga, K, Kanai, R, Sakamoto, O, Harata, K, Yamane, K. | | Deposit date: | 2003-09-01 | | Release date: | 2004-02-24 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Effects of Essential Carbohydrate/Aromatic Stacking Interaction with Tyr100 and Phe259 on Substrate Binding of Cyclodextrin Glycosyltransferase from Alkalophilic Bacillus sp. 1011
J.Biochem.(Tokyo), 134, 2003
|
|
1V3M
 
 | | Crystal structure of F283Y mutant cyclodextrin glycosyltransferase complexed with a pseudo-tetraose derived from acarbose | | Descriptor: | 4,6-dideoxy-alpha-D-xylo-hexopyranose-(1-4)-beta-D-galactopyranose, 6-AMINO-4-HYDROXYMETHYL-CYCLOHEX-4-ENE-1,2,3-TRIOL, CALCIUM ION, ... | | Authors: | Kanai, R, Haga, K, Akiba, T, Yamane, K, Harata, K. | | Deposit date: | 2003-11-03 | | Release date: | 2004-08-03 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Role of Phe283 in enzymatic reaction of cyclodextrin glycosyltransferase from alkalophilic Bacillus sp.1011: Substrate binding and arrangement of the catalytic site
PROTEIN SCI., 13, 2004
|
|
1V3K
 
 | | Crystal structure of F283Y mutant cyclodextrin glycosyltransferase | | Descriptor: | CALCIUM ION, Cyclomaltodextrin glucanotransferase | | Authors: | Kanai, R, Haga, K, Akiba, T, Yamane, K, Harata, K. | | Deposit date: | 2003-11-03 | | Release date: | 2004-08-03 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Role of Phe283 in enzymatic reaction of cyclodextrin glycosyltransferase from alkalophilic Bacillus sp.1011: substrate binding and arrangement of the catalytic site
PROTEIN SCI., 13, 2004
|
|
1V3J
 
 | | Crystal structure of F283L mutant cyclodextrin glycosyltransferase | | Descriptor: | CALCIUM ION, Cyclomaltodextrin glucanotransferase | | Authors: | Kanai, R, Haga, K, Akiba, T, Yamane, K, Harata, K. | | Deposit date: | 2003-11-03 | | Release date: | 2004-08-03 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Role of Phe283 in enzymatic reaction of cyclodextrin glycosyltransferase from alkalophilic Bacillus sp.1011: Substrate binding and arrangement of the catalytic site
PROTEIN SCI., 13, 2004
|
|
1V3L
 
 | | Crystal structure of F283L mutant cyclodextrin glycosyltransferase complexed with a pseudo-tetraose derived from acarbose | | Descriptor: | 4,6-dideoxy-alpha-D-xylo-hexopyranose-(1-4)-alpha-D-glucopyranose, 4,6-dideoxy-alpha-D-xylo-hexopyranose-(1-4)-beta-D-galactopyranose, 6-AMINO-4-HYDROXYMETHYL-CYCLOHEX-4-ENE-1,2,3-TRIOL, ... | | Authors: | Kanai, R, Haga, K, Akiba, T, Yamane, K, Harata, K. | | Deposit date: | 2003-11-03 | | Release date: | 2004-08-03 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Role of Phe283 in enzymatic reaction of cyclodextrin glycosyltransferase from alkalophilic Bacillus sp.1011: Substrate binding and arrangement of the catalytic site
PROTEIN SCI., 13, 2004
|
|
1VAH
 
 | | Crystal structure of the pig pancreatic-amylase complexed with r-nitrophenyl-a-D-maltoside | | Descriptor: | Alpha-amylase, pancreatic, CALCIUM ION, ... | | Authors: | Zhuo, H, Payan, F, Qian, M. | | Deposit date: | 2004-02-17 | | Release date: | 2005-04-26 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of the pig pancreatic alpha-amylase complexed with rho-nitrophenyl-alpha-D-maltoside-flexibility in the active site
Protein J., 23, 2004
|
|
