5MQF
 
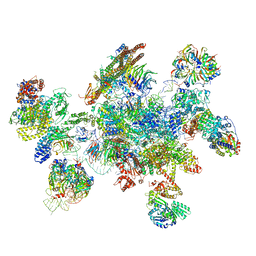 | | Cryo-EM structure of a human spliceosome activated for step 2 of splicing (C* complex) | | Descriptor: | 116 kDa U5 small nuclear ribonucleoprotein component, ATP-dependent RNA helicase DHX8, Cell division cycle 5-like protein, ... | | Authors: | Bertram, K, Hartmuth, K, Kastner, B. | | Deposit date: | 2016-12-20 | | Release date: | 2017-03-22 | | Last modified: | 2018-11-21 | | Method: | ELECTRON MICROSCOPY (5.9 Å) | | Cite: | Cryo-EM structure of a human spliceosome activated for step 2 of splicing.
Nature, 542, 2017
|
|
5MQ0
 
 | | Structure of a spliceosome remodeled for exon ligation | | Descriptor: | 3'-EXON OF UBC4 PRE-MRNA, BOUND BY PRP22 HELICASE, 5'-EXON OF UBC4 PRE-MRNA, ... | | Authors: | Fica, S.M, Oubridge, C, Galej, W.P, Wilkinson, M.E, Newman, A.J, Bai, X.-C, Nagai, K. | | Deposit date: | 2016-12-19 | | Release date: | 2017-01-18 | | Last modified: | 2020-10-07 | | Method: | ELECTRON MICROSCOPY (4.17 Å) | | Cite: | Structure of a spliceosome remodelled for exon ligation.
Nature, 542, 2017
|
|
1WCM
 
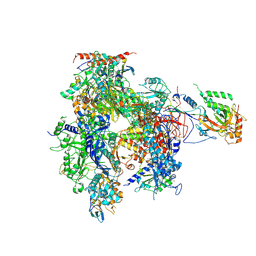 | | Complete 12-Subunit RNA Polymerase II at 3.8 Angstrom | | Descriptor: | DNA-DIRECTED RNA POLYMERASE II 13.6 KDA POLYPEPTIDE, DNA-DIRECTED RNA POLYMERASE II 14.2 KDA POLYPEPTIDE, DNA-DIRECTED RNA POLYMERASE II 19 KD POLYPEPTIDE, ... | | Authors: | Armache, K.-J, Mitterweger, S, Meinhart, A, Cramer, P. | | Deposit date: | 2004-11-17 | | Release date: | 2004-12-14 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | Structures of Complete RNA Polymerase II and its Subcomplex,Rpb4/7
J.Biol.Chem., 280, 2005
|
|
2PMZ
 
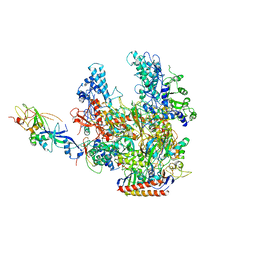 | | Archaeal RNA polymerase from Sulfolobus solfataricus | | Descriptor: | DNA-directed RNA polymerase subunit A, DNA-directed RNA polymerase subunit A", DNA-directed RNA polymerase subunit B, ... | | Authors: | Murakami, K.S. | | Deposit date: | 2007-04-23 | | Release date: | 2008-02-12 | | Last modified: | 2017-10-18 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | The X-ray crystal structure of RNA polymerase from Archaea
Nature, 451, 2008
|
|
5OIK
 
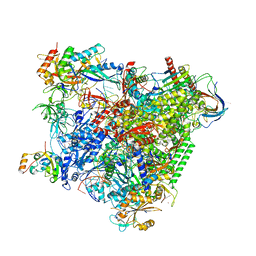 | | Structure of an RNA polymerase II-DSIF transcription elongation complex | | Descriptor: | DNA (43-MER), DNA-directed RNA polymerase II subunit RPB1, DNA-directed RNA polymerase II subunit RPB11, ... | | Authors: | Bernecky, C, Plitzko, J.M, Cramer, P. | | Deposit date: | 2017-07-18 | | Release date: | 2017-09-13 | | Last modified: | 2019-12-11 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structure of a transcribing RNA polymerase II-DSIF complex reveals a multidentate DNA-RNA clamp.
Nat. Struct. Mol. Biol., 24, 2017
|
|
5OQM
 
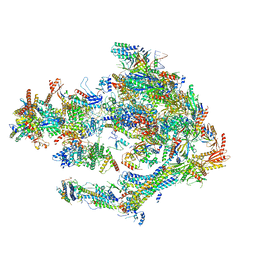 | | STRUCTURE OF YEAST TRANSCRIPTION PRE-INITIATION COMPLEX WITH TFIIH AND CORE MEDIATOR | | Descriptor: | DNA-directed RNA polymerase II subunit RPB1, DNA-directed RNA polymerase II subunit RPB11, DNA-directed RNA polymerase II subunit RPB2, ... | | Authors: | Schilbach, S, Hantsche, M, Tegunov, D, Dienemann, C, Wigge, C, Urlaub, H, Cramer, P. | | Deposit date: | 2017-08-13 | | Release date: | 2018-05-09 | | Last modified: | 2018-11-21 | | Method: | ELECTRON MICROSCOPY (5.8 Å) | | Cite: | Structures of transcription pre-initiation complex with TFIIH and Mediator.
Nature, 551, 2017
|
|
2R7Z
 
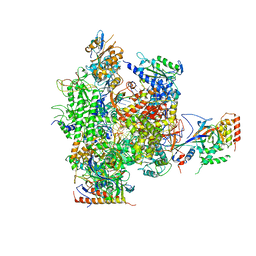 | | Cisplatin lesion containing RNA polymerase II elongation complex | | Descriptor: | 5'-D(*CP*AP*AP*GP*TP*AP*G)-3', 5'-D(*TP*AP*CP*TP*TP*GUP*CP*CP*CP*TP*CP*CP*TP*CP*AP*T)-3', 5'-R(*UP*UP*UP*GP*AP*GP*GP*AP*GP*G)-3', ... | | Authors: | Damsma, G.E, Alt, A, Brueckner, F, Carell, T, Cramer, P. | | Deposit date: | 2007-09-10 | | Release date: | 2007-11-20 | | Last modified: | 2017-10-25 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | Mechanism of transcriptional stalling at cisplatin-damaged DNA.
Nat.Struct.Mol.Biol., 14, 2007
|
|
2R92
 
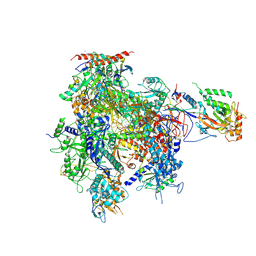 | | Elongation complex of RNA polymerase II with artificial RdRP scaffold | | Descriptor: | DNA-directed RNA polymerase II subunit RPB1, DNA-directed RNA polymerase II subunit RPB11, DNA-directed RNA polymerase II subunit RPB2, ... | | Authors: | Lehmann, E, Brueckner, F, Cramer, P. | | Deposit date: | 2007-09-12 | | Release date: | 2007-11-27 | | Last modified: | 2017-10-25 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | Molecular basis of RNA-dependent RNA polymerase II activity.
Nature, 450, 2007
|
|
1Y1Y
 
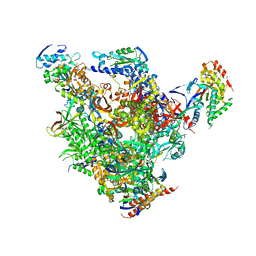 | | RNA Polymerase II-TFIIS-DNA/RNA complex | | Descriptor: | 5'-D(P*TP*AP*CP*GP*CP*CP*T)-3', 5'-R(P*AP*GP*GP*C)-3', DNA-directed RNA polymerase II 13.6 kDa polypeptide, ... | | Authors: | Cramer, P, Kettenberger, H, Armache, K.-J. | | Deposit date: | 2004-11-19 | | Release date: | 2004-12-28 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (4 Å) | | Cite: | Complete RNA polymerase II elongation complex structure and its interactions with NTP and TFIIS.
Mol.Cell, 16, 2004
|
|
1Y77
 
 | | Complete RNA Polymerase II elongation complex with substrate analogue GMPCPP | | Descriptor: | 5'-D(*AP*AP*GP*TP*AP*CP*T)-3', 5'-D(P*AP*GP*TP*AP*CP*TP*TP*AP*CP*T*CP*GP*CP*CP*TP*GP*GP*TP*CP*TP*G)-3', 5'-R(*AP*AP*GP*AP*CP*CP*AP*GP*GP*C)-3', ... | | Authors: | Kettenberger, H, Armache, K.-J, Cramer, P. | | Deposit date: | 2004-12-08 | | Release date: | 2005-01-04 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (4.5 Å) | | Cite: | Complete RNA Polymerase II Elongation Complex Structure and Its Interactions with NTP and TFIIS
Mol.Cell, 16, 2004
|
|
1Y1W
 
 | | Complete RNA Polymerase II elongation complex | | Descriptor: | 5'-D(*AP*AP*GP*TP*AP*CP*T)-3', 5'-D(P*AP*GP*TP*AP*CP*TP*TP*AP*CP*GP*CP*CP*TP*GP*GP*TP*CP*AP*T)-3', 5'-R(*AP*AP*GP*AP*CP*CP*AP*GP*GP*C)-3', ... | | Authors: | Cramer, P, Kettenberger, H, Armache, K.-J. | | Deposit date: | 2004-11-19 | | Release date: | 2005-01-04 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (4 Å) | | Cite: | Complete RNA Polymerase II Elongation Complex Structure and Its Interactions with NTP and TFIIS
Mol.Cell, 16, 2004
|
|
2R93
 
 | | Elongation complex of RNA polymerase II with a hepatitis delta virus-derived RNA stem loop | | Descriptor: | DNA-directed RNA polymerase II subunit RPB1, DNA-directed RNA polymerase II subunit RPB11, DNA-directed RNA polymerase II subunit RPB2, ... | | Authors: | Lehmann, E, Brueckner, F, Cramer, P. | | Deposit date: | 2007-09-12 | | Release date: | 2007-11-27 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (4 Å) | | Cite: | Molecular basis of RNA-dependent RNA polymerase II activity.
Nature, 450, 2007
|
|
1Y1V
 
 | | Refined RNA Polymerase II-TFIIS complex | | Descriptor: | DNA-directed RNA polymerase II 13.6 kDa polypeptide, DNA-directed RNA polymerase II 140 kDa polypeptide, DNA-directed RNA polymerase II 19 kDa polypeptide, ... | | Authors: | Kettenberger, H, Armache, K.-J, Cramer, P. | | Deposit date: | 2004-11-19 | | Release date: | 2004-12-28 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | Complete RNA polymerase II elongation complex structure and its interactions with NTP and TFIIS.
Mol.Cell, 16, 2004
|
|
2WB1
 
 | | The complete structure of the archaeal 13-subunit DNA-directed RNA Polymerase | | Descriptor: | DNA-DIRECTED RNA POLYMERASE RPO10 SUBUNIT, DNA-DIRECTED RNA POLYMERASE RPO11 SUBUNIT, DNA-DIRECTED RNA POLYMERASE RPO12 SUBUNIT, ... | | Authors: | Korkhin, Y, Unligil, U.M, Littlefield, O, Nelson, P.J, Stuart, D.I, Sigler, P.B, Bell, S.D, Abrescia, N.G.A. | | Deposit date: | 2009-02-19 | | Release date: | 2009-05-19 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3.52 Å) | | Cite: | Evolution of Complex RNA Polymerase: The Complete Archaeal RNA Polymerase Structure
Plos Biol., 7, 2009
|
|
1NT9
 
 | | Complete 12-subunit RNA polymerase II | | Descriptor: | DNA-DIRECTED RNA POLYMERASE II 13.6 KD POLYPEPTIDE, DNA-DIRECTED RNA POLYMERASE II 19 KD POLYPEPTIDE, DNA-DIRECTED RNA POLYMERASE II LARGEST SUBUNIT, ... | | Authors: | Armache, K.-J, Kettenberger, H, Cramer, P. | | Deposit date: | 2003-01-29 | | Release date: | 2003-04-22 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (4.2 Å) | | Cite: | Architecture of initiation-competent 12-subunit RNA polymerase II
Proc.Natl.Acad.Sci.USA, 100, 2003
|
|
1PQV
 
 | | RNA polymerase II-TFIIS complex | | Descriptor: | DNA-directed RNA polymerase II 13.6 kDa polypeptide, DNA-directed RNA polymerase II 14.2 kDa polypeptide, DNA-directed RNA polymerase II 140 kDa polypeptide, ... | | Authors: | Kettenberger, H, Armache, K.-J, Cramer, P. | | Deposit date: | 2003-06-19 | | Release date: | 2003-08-19 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | Architecture of the RNA Polymerase II-TFIIS Complex and Implications for mRNA Cleavage
Cell(Cambridge,Mass.), 114, 2003
|
|
7QGH
 
 | | Structure of the E. coli disome - collided 70S ribosome | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S1, ... | | Authors: | Kratzat, H, Buschauer, R, Berninghausen, O, Beckmann, R. | | Deposit date: | 2021-12-08 | | Release date: | 2022-03-16 | | Last modified: | 2022-03-30 | | Method: | ELECTRON MICROSCOPY (4.48 Å) | | Cite: | Ribosome collisions induce mRNA cleavage and ribosome rescue in bacteria.
Nature, 603, 2022
|
|
7PY5
 
 | | CryoEM structure of E.coli RNA polymerase elongation complex bound to NusA and NusG (the consensus NusA-NusG-EC) | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | Zhu, C, Guo, X, Weixlbaumer, A. | | Deposit date: | 2021-10-09 | | Release date: | 2022-03-23 | | Last modified: | 2022-10-05 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Transcription factors modulate RNA polymerase conformational equilibrium.
Nat Commun, 13, 2022
|
|
7PY3
 
 | | CryoEM structure of E.coli RNA polymerase elongation complex bound to NusA (the consensus NusA-EC) | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | Zhu, C, Guo, X, Weixlbaumer, A. | | Deposit date: | 2021-10-08 | | Release date: | 2022-03-23 | | Last modified: | 2022-10-05 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Transcription factors modulate RNA polymerase conformational equilibrium.
Nat Commun, 13, 2022
|
|
7PY6
 
 | | CryoEM structure of E.coli RNA polymerase elongation complex bound to NusA and NusG (NusA and NusG elongation complex in less-swiveled conformation) | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | Zhu, C, Guo, X, Weixlbaumer, A. | | Deposit date: | 2021-10-09 | | Release date: | 2022-03-23 | | Last modified: | 2022-10-05 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Transcription factors modulate RNA polymerase conformational equilibrium.
Nat Commun, 13, 2022
|
|
7PY7
 
 | | CryoEM structure of E.coli RNA polymerase elongation complex bound to NusA and NusG (NusA and NusG elongation complex in more-swiveled conformation) | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | Zhu, C, Guo, X, Weixlbaumer, A. | | Deposit date: | 2021-10-09 | | Release date: | 2022-03-23 | | Last modified: | 2022-10-05 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Transcription factors modulate RNA polymerase conformational equilibrium.
Nat Commun, 13, 2022
|
|
7PYK
 
 | | CryoEM structure of E.coli RNA polymerase elongation complex bound to NusA (NusA elongation complex in more-swiveled conformation) | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | Zhu, C, Guo, X, Weixlbaumer, A. | | Deposit date: | 2021-10-10 | | Release date: | 2022-03-23 | | Last modified: | 2022-10-05 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Transcription factors modulate RNA polymerase conformational equilibrium.
Nat Commun, 13, 2022
|
|
7PYJ
 
 | | CryoEM structure of E.coli RNA polymerase elongation complex bound to NusA (NusA elongation complex in less-swiveled conformation) | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | Zhu, C, Guo, X, Weixlbaumer, A. | | Deposit date: | 2021-10-10 | | Release date: | 2022-03-23 | | Last modified: | 2022-10-05 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Transcription factors modulate RNA polymerase conformational equilibrium.
Nat Commun, 13, 2022
|
|
7QP7
 
 | | Structure of the human 48S initiation complex in closed state (h48S AUG closed) | | Descriptor: | 18S rRNA, 40S ribosomal protein S10, 40S ribosomal protein S11, ... | | Authors: | Yi, S.-H, Petrychenko, V, Schliep, J.E, Goyal, A, Linden, A, Chari, A, Urlaub, H, Stark, H, Rodnina, M.V, Adio, S, Fischer, N. | | Deposit date: | 2022-01-03 | | Release date: | 2022-05-11 | | Last modified: | 2022-06-01 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Conformational rearrangements upon start codon recognition in human 48S translation initiation complex.
Nucleic Acids Res., 50, 2022
|
|
7QP6
 
 | | Structure of the human 48S initiation complex in open state (h48S AUG open) | | Descriptor: | 18S rRNA, 40S ribosomal protein S10, 40S ribosomal protein S11, ... | | Authors: | Yi, S.-H, Petrychenko, V, Schliep, J.E, Goyal, A, Linden, A, Chari, A, Urlaub, H, Stark, H, Rodnina, M.V, Adio, S, Fischer, N. | | Deposit date: | 2022-01-03 | | Release date: | 2022-05-11 | | Last modified: | 2022-06-01 | | Method: | ELECTRON MICROSCOPY (4.7 Å) | | Cite: | Conformational rearrangements upon start codon recognition in human 48S translation initiation complex.
Nucleic Acids Res., 50, 2022
|
|
