3LL8
 
 | |
3QFK
 
 | | 2.05 Angstrom Crystal Structure of Putative 5'-Nucleotidase from Staphylococcus aureus in complex with alpha-ketoglutarate | | Descriptor: | 1,2-ETHANEDIOL, 2-OXOGLUTARIC ACID, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Minasov, G, Wawrzak, Z, Krishna, S.N, Halavaty, A, Shuvalova, L, Dubrovska, I, Winsor, J, Kiryukhina, O, Bagnoli, F, Falugi, F, Bottomley, M, Grandi, G, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-01-21 | | Release date: | 2011-02-09 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | 2.05 Angstrom Crystal Structure of Putative 5'-Nucleotidase from Staphylococcus aureus in complex with alpha-ketoglutarate.
TO BE PUBLISHED
|
|
3P71
 
 | | Crystal structure of the complex of LCMT-1 and PP2A | | Descriptor: | 5'-{[(3S)-3-amino-3-carboxypropyl](ethyl)amino}-5'-deoxyadenosine, DI(HYDROXYETHYL)ETHER, Leucine carboxyl methyltransferase 1, ... | | Authors: | Xing, Y, Stanevich, V, Satyshur, K.A, Jiang, L. | | Deposit date: | 2010-10-11 | | Release date: | 2011-02-16 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The Structural Basis for Tight Control of PP2A Methylation and Function by LCMT-1.
Mol.Cell, 41, 2011
|
|
2XMO
 
 | | The crystal structure of Lmo2642 | | Descriptor: | CALCIUM ION, FE (III) ION, LMO2642 PROTEIN, ... | | Authors: | Jeong, J.H, Kim, Y.G. | | Deposit date: | 2010-07-28 | | Release date: | 2011-02-23 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural and Functional Analysis of the Lmo2642 Cyclic Nucleotide Phosphodiesterase from Listeria Monocytogenes.
Proteins, 79, 2011
|
|
3QG5
 
 | |
3AV0
 
 | | Crystal structure of Mre11-Rad50 bound to ATP S | | Descriptor: | DNA double-strand break repair protein mre11, DNA double-strand break repair rad50 ATPase, GLYCEROL, ... | | Authors: | Lim, H.S, Kim, J.S, Cho, Y. | | Deposit date: | 2011-02-18 | | Release date: | 2011-05-25 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Crystal Structure of the Mre11-Rad50-ATP S Complex: Understanding the Interplay between Mre11 and Rad50
To be Published
|
|
3AUZ
 
 | | Crystal structure of Mre11 with manganese | | Descriptor: | DNA double-strand break repair protein mre11, GLYCEROL, MANGANESE (II) ION | | Authors: | Park, Y.B, Cho, Y. | | Deposit date: | 2011-02-18 | | Release date: | 2011-05-25 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.206 Å) | | Cite: | Crystal Structure of the Mre11-Rad50-ATP S Complex: Understanding the Interplay between Mre11 and Rad50
To be Published
|
|
3RL5
 
 | | Rat metallophosphodiesterase MPPED2 H67R Mutant | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, Metallophosphoesterase MPPED2, ... | | Authors: | Podobnik, M, Dermol, U. | | Deposit date: | 2011-04-19 | | Release date: | 2011-08-10 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.26 Å) | | Cite: | Unique utilization of a phosphoprotein phosphatase fold by a mammalian phosphodiesterase associated with WAGR syndrome.
J.Mol.Biol., 412, 2011
|
|
3RL4
 
 | | Rat metallophosphodiesterase MPPED2 G252H Mutant | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, GUANOSINE-5'-MONOPHOSPHATE, ... | | Authors: | Podobnik, M, Dermol, U. | | Deposit date: | 2011-04-19 | | Release date: | 2011-08-10 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.29 Å) | | Cite: | Unique utilization of a phosphoprotein phosphatase fold by a mammalian phosphodiesterase associated with WAGR syndrome.
J.Mol.Biol., 412, 2011
|
|
3RL3
 
 | | Rat metallophosphodiesterase MPPED2 | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, GUANOSINE-5'-MONOPHOSPHATE, ... | | Authors: | Podobnik, M, Dermol, U. | | Deposit date: | 2011-04-19 | | Release date: | 2011-08-10 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Unique utilization of a phosphoprotein phosphatase fold by a mammalian phosphodiesterase associated with WAGR syndrome.
J.Mol.Biol., 412, 2011
|
|
3THN
 
 | | Crystal structure of Mre11 core with manganese | | Descriptor: | Exonuclease, putative, MANGANESE (II) ION, ... | | Authors: | Moeckel, C, Lammens, K. | | Deposit date: | 2011-08-19 | | Release date: | 2011-10-12 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.811 Å) | | Cite: | ATP driven structural changes of the bacterial Mre11:Rad50 catalytic head complex.
Nucleic Acids Res., 40, 2012
|
|
3THO
 
 | |
3T1I
 
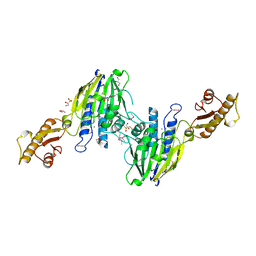 | | Crystal Structure of Human Mre11: Understanding Tumorigenic Mutations | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, Double-strand break repair protein MRE11A, GLYCEROL, ... | | Authors: | Park, Y.B, Chae, J, Kim, Y, Cho, Y. | | Deposit date: | 2011-07-22 | | Release date: | 2011-11-30 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structure of human mre11: understanding tumorigenic mutations
Structure, 19, 2011
|
|
3ZU0
 
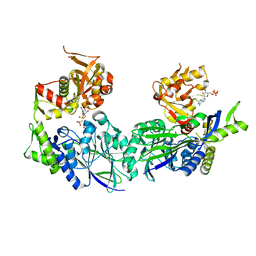 | | Structure of Haemophilus influenzae NAD nucleotidase (NadN) | | Descriptor: | NAD NUCLEOTIDASE, PHOSPHATE ION, PHOSPHOMETHYLPHOSPHONIC ACID ADENOSYL ESTER, ... | | Authors: | Garavaglia, S, Bruzzone, S, Cassani, C, Canella, L, Allegrone, G, Sturla, L, Mannino, E, Millo, E, De Flora, A, Rizzi, M. | | Deposit date: | 2011-07-13 | | Release date: | 2011-12-14 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.001 Å) | | Cite: | The High-Resolution Crystal Structure of Periplasmic Haemophilus Influenzae Nad Nucleotidase Reveals a Novel Enzymatic Function of Human Cd73 Related to Nad Metabolism.
Biochem.J., 441, 2012
|
|
3ZTV
 
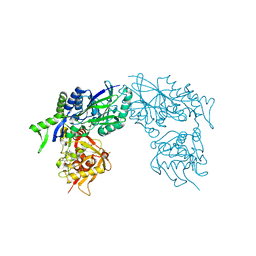 | | Structure of Haemophilus influenzae NAD nucleotidase (NadN) | | Descriptor: | ADENOSINE, GLYCEROL, NAD NUCLEOTIDASE, ... | | Authors: | Garavaglia, S, Bruzzone, S, Cassani, C, Canella, L, Allegrone, G, Sturla, L, Mannino, E, Millo, E, De Flora, A, Rizzi, M. | | Deposit date: | 2011-07-12 | | Release date: | 2011-12-14 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | The High-Resolution Crystal Structure of Periplasmic Haemophilus Influenzae Nad Nucleotidase Reveals a Novel Enzymatic Function of Human Cd73 Related to Nad Metabolism.
Biochem.J., 441, 2012
|
|
3TGH
 
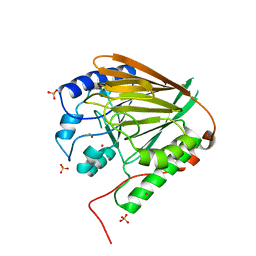 | | GAP50 the anchor in the inner membrane complex of Plasmodium | | Descriptor: | COBALT (II) ION, DIMETHYL SULFOXIDE, Glideosome-associated protein 50, ... | | Authors: | Bosch, J, Paige, M.H, Vaidya, A, Bergman, L, Hol, W.G.J. | | Deposit date: | 2011-08-17 | | Release date: | 2012-03-07 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of GAP50, the anchor of the invasion machinery in the inner membrane complex of Plasmodium falciparum.
J.Struct.Biol., 178, 2012
|
|
4FCX
 
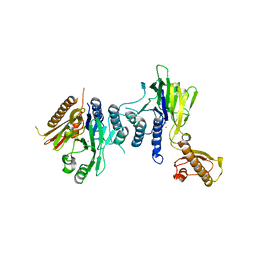 | | S.pombe Mre11 apoenzym | | Descriptor: | DNA repair protein rad32, MANGANESE (II) ION | | Authors: | Schiller, C.B, Lammens, K, Hopfner, K.P. | | Deposit date: | 2012-05-25 | | Release date: | 2012-06-20 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure of Mre11-Nbs1 complex yields insights into ataxia-telangiectasia-like disease mutations and DNA damage signaling.
Nat.Struct.Mol.Biol., 19, 2012
|
|
4FBW
 
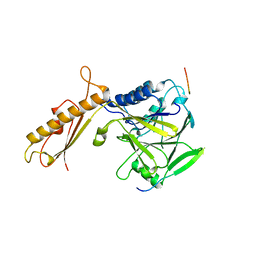 | |
4FBK
 
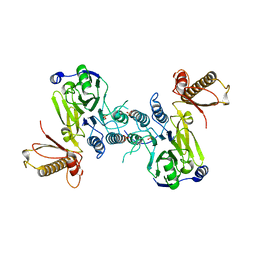 | | Crystal structure of a covalently fused Nbs1-Mre11 complex with one manganese ion per active site | | Descriptor: | DNA repair and telomere maintenance protein nbs1,DNA repair protein rad32 CHIMERIC PROTEIN, MANGANESE (II) ION, SULFATE ION | | Authors: | Schiller, C.B, Lammens, K, Hopfner, K.P. | | Deposit date: | 2012-05-23 | | Release date: | 2012-06-20 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.379 Å) | | Cite: | Structure of Mre11-Nbs1 complex yields insights into ataxia-telangiectasia-like disease mutations and DNA damage signaling.
Nat.Struct.Mol.Biol., 19, 2012
|
|
4FBQ
 
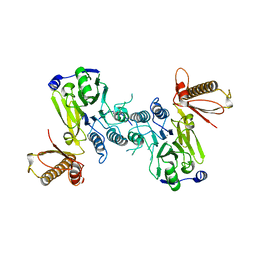 | |
4G9J
 
 | | Protein Ser/Thr phosphatase-1 in complex with cell-permeable peptide | | Descriptor: | MANGANESE (II) ION, Serine/threonine-protein phosphatase PP1-alpha catalytic subunit, synthetic peptide | | Authors: | Sukackaite, R, Chatterjee, J, Beullens, M, Bollen, M, Koehn, M, Hart, D.J. | | Deposit date: | 2012-07-24 | | Release date: | 2012-09-19 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Development of a Peptide that Selectively Activates Protein Phosphatase-1 in Living Cells.
Angew.Chem.Int.Ed.Engl., 51, 2012
|
|
4DT2
 
 | | Crystal structure of red kidney bean purple acid phosphatase in complex with Maybridge fragment CC27209 | | Descriptor: | (2,2-dimethyl-2,3-dihydro-1-benzofuran-7-yl)methanol, 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Feder, D, Hussein, W.M, Clayton, D.J, Kan, M, Schenk, G, McGeary, R.P, Guddat, L.W. | | Deposit date: | 2012-02-20 | | Release date: | 2012-09-19 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Identification of purple acid phosphatase inhibitors by fragment-based screening: promising new leads for osteoporosis therapeutics.
Chem.Biol.Drug Des., 80, 2012
|
|
4DSY
 
 | | Crystal structure of red kidney bean purple acid phosphatase in complex with Maybridge fragment CC24201 | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 5-phenylpyridine-3-carboxylic acid, ... | | Authors: | Feder, D, Hussein, W.M, Clayton, D.J, Kan, M, Schenk, G, McGeary, R.P, Guddat, L.W. | | Deposit date: | 2012-02-20 | | Release date: | 2012-09-19 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Identification of purple acid phosphatase inhibitors by fragment-based screening: promising new leads for osteoporosis therapeutics.
Chem.Biol.Drug Des., 80, 2012
|
|
4H1S
 
 | | Crystal Structure of a Truncated Soluble form of Human CD73 with Ecto-5'-Nucleotidase activity | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 5'-nucleotidase, PHOSPHATE ION, ... | | Authors: | Heuts, D.P, Weissenborn, M.J, Olkhov, R.V, Shaw, A.M, Levy, C.W, Scrutton, N.S. | | Deposit date: | 2012-09-11 | | Release date: | 2012-10-03 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of a soluble form of human CD73 with ecto-5'-nucleotidase activity.
Chembiochem, 13, 2012
|
|
4HD0
 
 | | Mre11 ATLD17/18 mutation retains Tel1/ATM activity but blocks DNA double-strand break repair | | Descriptor: | DNA double-strand break repair protein Mre11, MANGANESE (II) ION | | Authors: | Limbo, O, Moiani, D, Kertokalio, A, Wyman, C, Tainer, J.A, Russell, P. | | Deposit date: | 2012-10-01 | | Release date: | 2012-10-31 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Mre11 ATLD17/18 mutation retains Tel1/ATM activity but blocks DNA double-strand break repair.
Nucleic Acids Res., 40, 2012
|
|
