6GXF
 
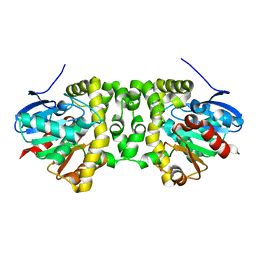 | | The hit-and-return system enables efficient time-resolved serial synchrotron crystallography: RADDAM1 | | Descriptor: | Fluoroacetate dehalogenase | | Authors: | Schulz, E.C, Mehrabi, P, Mueller-Werkmeister, H, Tellkamp, F, Stuart, W, Persch, E, De Gasparo, R, Diederich, F, Pai, E.F, Miller, R.J.D. | | Deposit date: | 2018-06-27 | | Release date: | 2018-10-17 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The hit-and-return system enables efficient time-resolved serial synchrotron crystallography.
Nat. Methods, 15, 2018
|
|
4KAC
 
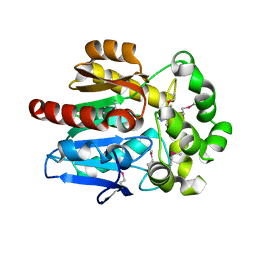 | | X-Ray Structure of the complex HaloTag2 with HALTS. Northeast Structural Genomics Consortium (NESG) Target OR150. | | Descriptor: | 2-{2-[2-(2-{2-[2-(2-ETHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, AMMONIUM ION, Haloalkane dehalogenase, ... | | Authors: | Kuzin, A, Lew, S, Neklesa, T.K, Noblin, D, Seetharaman, J, Maglaqui, M, Xiao, R, Kohan, E, Wang, H, Everett, J.K, Acton, T.B, Kornhaber, G, Montelione, G.T, Crews, C.M, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2013-04-22 | | Release date: | 2013-06-05 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.223 Å) | | Cite: | Northeast Structural Genomics Consortium Target OR150
To be Published
|
|
4KAJ
 
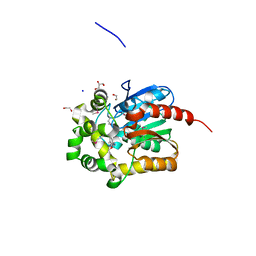 | | X-Ray Structure of the complex of Haloalkane dehalogenase HaloTag7 with HALTS, Northeast Structural Genomics Consortium (NESG) Target OR151 | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, MANGANESE (II) ION, ... | | Authors: | Kuzin, A, Lew, S, Neklesa, T.K, Noblin, D, Seetharaman, J, Maglaqui, M, Xiao, R, Kohan, E, Wang, H, Everett, J.K, Acton, T.B, Kornhaber, G, Montelione, G.T, Crews, C.M, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2013-04-22 | | Release date: | 2013-06-05 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.948 Å) | | Cite: | Northeast Structural Genomics Consortium Target OR151
To be Published
|
|
7AL5
 
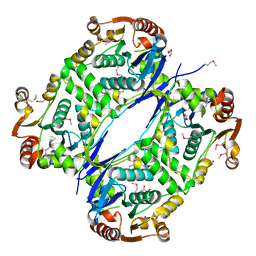 | |
7AL6
 
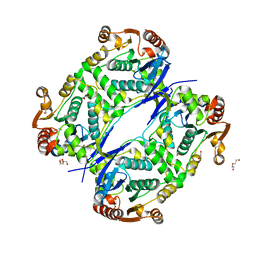 | |
7M7C
 
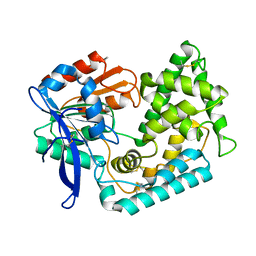 | | Crystal Structure of Hip1 (Rv2224c) mutant - T466A/S228DHA (dehydroalanine) | | Descriptor: | Carboxylesterase A | | Authors: | Naffin-Olivos, J.L, Daab, A, Goldfarb, N.E, Doran, M.H, Baikovitz, J, Liu, D, Sun, S, White, A, Dunn, B.M, Rengarajan, J, Petsko, G.A, Ringe, D. | | Deposit date: | 2021-03-27 | | Release date: | 2022-03-30 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Inhibitors and Inactivators of Mycobacterium tuberculosis serine protease Hip1 (Rv2224c)
To Be Published
|
|
7AVR
 
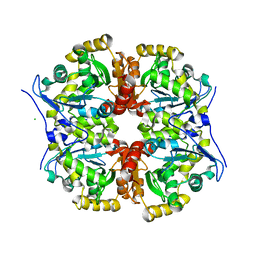 | | The tetrameric structure of haloalkane dehalogenase DpaA from Paraglaciecola agarilytica NO2 | | Descriptor: | CHLORIDE ION, Haloalkane dehalogenase 1 | | Authors: | Mazur, A, Kolenko, P, Prudnikova, T, Grinkevich, P, Kuta Smatanova, I. | | Deposit date: | 2020-11-05 | | Release date: | 2021-03-17 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The tetrameric structure of the novel haloalkane dehalogenase DpaA from Paraglaciecola agarilytica NO2.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
4L9A
 
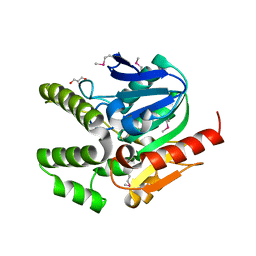 | |
4LIP
 
 | |
7COF
 
 | |
7NFZ
 
 | |
7CLZ
 
 | |
7COG
 
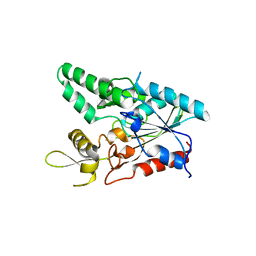 | |
7OND
 
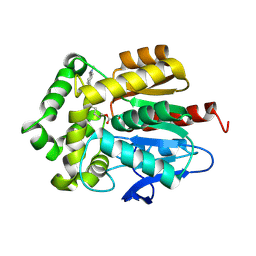 | |
7OO4
 
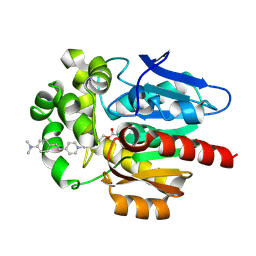 | |
7C4D
 
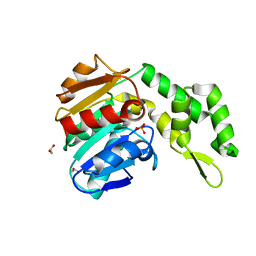 | | Marine microorganism esterase | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, Putative esterase, ... | | Authors: | Zhu, C.H, Wu, Y.K, Isupov, M.N. | | Deposit date: | 2020-05-16 | | Release date: | 2021-05-26 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Structural Insights into a Novel Esterase from the East Pacific Rise and Its Improved Thermostability by a Semirational Design.
J.Agric.Food Chem., 69, 2021
|
|
7O3O
 
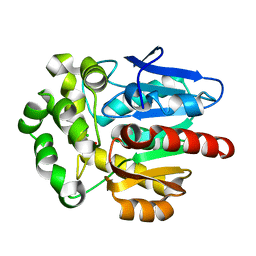 | | Structure of haloalkane dehalogenase mutant DhaA80(T148L, G171Q, A172V, C176F) from Rhodococcus rhodochrous with ionic liquid | | Descriptor: | CHLORIDE ION, ETHANOLAMINE, Haloalkane dehalogenase | | Authors: | Shaposhnikova, A, Prudnikova, T, Kuta Smatanova, I. | | Deposit date: | 2021-04-02 | | Release date: | 2021-09-08 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Stabilization of Haloalkane Dehalogenase Structure by Interfacial Interaction with Ionic Liquids
Crystals, 11, 2021
|
|
7O8B
 
 | |
7CG2
 
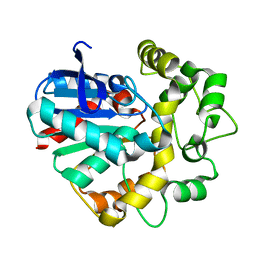 | | Vigna radiata Epoxide hydrolase mutant | | Descriptor: | Epoxide hydrolase | | Authors: | Li, F.L, Yu, H.L, Xu, J.H. | | Deposit date: | 2020-06-30 | | Release date: | 2021-07-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.001 Å) | | Cite: | Reprogramming Epoxide Hydrolase to Improve Enantioconvergence in Hydrolysis of Styrene Oxide Scaffolds
Adv.Synth.Catal., 362, 2021
|
|
7CG6
 
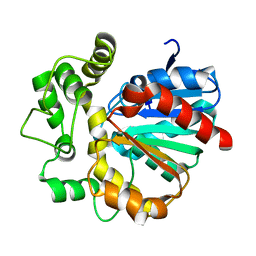 | | Vigna radiata Epoxide hydrolase mutant M263Q | | Descriptor: | Epoxide hydrolase | | Authors: | Li, F.L, Yu, H.L, Xu, J.H. | | Deposit date: | 2020-06-30 | | Release date: | 2021-07-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Reprogramming Epoxide Hydrolase to Improve Enantioconvergence in Hydrolysis of Styrene Oxide Scaffolds
Adv.Synth.Catal., 362, 2021
|
|
7D78
 
 | | The structure of thioesterase DcsB | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, DltD domain-containing protein, ... | | Authors: | Tang, Y, Zhou, J.H, Wang, G.Q. | | Deposit date: | 2020-10-03 | | Release date: | 2021-01-27 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.96543121 Å) | | Cite: | A Polyketide Cyclase That Forms Medium-Ring Lactones.
J.Am.Chem.Soc., 143, 2021
|
|
7D79
 
 | | The structure of DcsB complex with its substrate analogue | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, DltD domain-containing protein, ... | | Authors: | Tang, Y, Zhou, J.H, Wang, G.Q. | | Deposit date: | 2020-10-03 | | Release date: | 2021-01-27 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.10411429 Å) | | Cite: | A Polyketide Cyclase That Forms Medium-Ring Lactones.
J.Am.Chem.Soc., 143, 2021
|
|
7OKZ
 
 | | CRYSTAL STRUCTURE OF THE COFACTOR-DEVOID 1-H-3-HYDROXY-4- OXOQUINALDINE 2,4-DIOXYGENASE (HOD) CATALYTICALLY INACTIVE H251A VARIANT COMPLEXED WITH 2-METHYL- QUINOLIN-4(1H)-ONE UNDER HYPEROXIC CONDITIONS | | Descriptor: | 1H-3-hydroxy-4-oxoquinaldine 2,4-dioxygenase, 2-methyl-quinolin-4(1H)-one, D(-)-TARTARIC ACID, ... | | Authors: | Bui, S, Steiner, R.A. | | Deposit date: | 2021-05-18 | | Release date: | 2022-06-01 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.101 Å) | | Cite: | Evolutionary adaptation from hydrolytic to oxygenolytic catalysis at the alpha / beta-hydrolase fold.
Chem Sci, 14, 2023
|
|
7OMD
 
 | | Crystal structure of azacoelenterazine-bound Renilla reniformis luciferase variant RLuc8-D162A | | Descriptor: | 6-(4-hydroxyphenyl)-2-[(4-hydroxyphenyl)methyl]-8-(phenylmethyl)-[1,2,4]triazolo[4,3-a]pyrazin-3-one, CHLORIDE ION, Coelenterazine h 2-monooxygenase, ... | | Authors: | Schenkmayerova, A, Janin, Y.L, Marek, M. | | Deposit date: | 2021-05-21 | | Release date: | 2022-06-01 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.601 Å) | | Cite: | Catalytic mechanism for Renilla-type luciferases
Nat Catal, 2023
|
|
7OJM
 
 | | CRYSTAL STRUCTURE OF THE COFACTOR-DEVOID 1-H-3-HYDROXY-4- OXOQUINALDINE 2,4-DIOXYGENASE (HOD) CATALYTICALLY INACTIVE H251A VARIANT COMPLEXED WITH 2-METHYL-QUINOLIN-4(1H)-ONE UNDER NORMOXIC CONDITIONS | | Descriptor: | 1H-3-hydroxy-4-oxoquinaldine 2,4-dioxygenase, 2-methyl-quinolin-4(1H)-one, GLYCEROL, ... | | Authors: | Bui, S, Steiner, R.A. | | Deposit date: | 2021-05-16 | | Release date: | 2022-06-01 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.001 Å) | | Cite: | Evolutionary adaptation from hydrolytic to oxygenolytic catalysis at the alpha / beta-hydrolase fold.
Chem Sci, 14, 2023
|
|
