2PRU
 
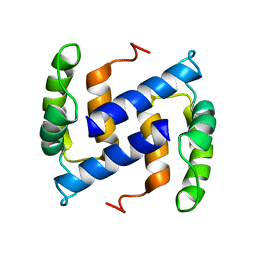 | | NMR Structure of Human apoS100B at 10C | | Descriptor: | Protein S100-B | | Authors: | Malik, S, Shaw, G.S, Revington, M. | | Deposit date: | 2007-05-04 | | Release date: | 2008-04-15 | | Last modified: | 2022-03-16 | | Method: | SOLUTION NMR | | Cite: | Analysis of the structure of human apo-S100B at low temperature indicates a unimodal conformational distribution is adopted by calcium-free S100 proteins.
Proteins, 73, 2008
|
|
2JPT
 
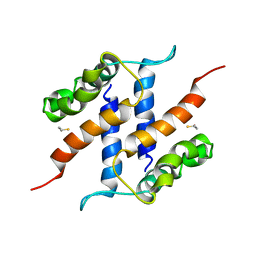 | |
2JU0
 
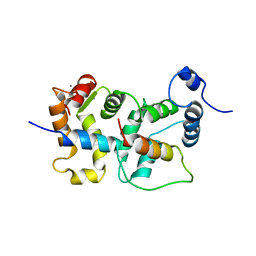 | | Structure of Yeast Frequenin bound to PdtIns 4-kinase | | Descriptor: | CALCIUM ION, Calcium-binding protein NCS-1, Phosphatidylinositol 4-kinase PIK1 | | Authors: | Ames, J. | | Deposit date: | 2007-08-11 | | Release date: | 2007-08-28 | | Last modified: | 2022-03-16 | | Method: | SOLUTION NMR | | Cite: | Structural insights into activation of phosphatidylinositol 4-kinase (Pik1) by yeast frequenin (Frq1).
J.Biol.Chem., 282, 2007
|
|
2R2I
 
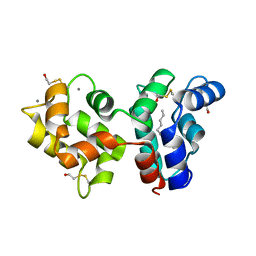 | | Myristoylated Guanylate Cyclase Activating Protein-1 with Calcium Bound | | Descriptor: | BETA-MERCAPTOETHANOL, CALCIUM ION, Guanylyl cyclase-activating protein 1, ... | | Authors: | Stephen, R. | | Deposit date: | 2007-08-25 | | Release date: | 2007-12-11 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Stabilizing function for myristoyl group revealed by the crystal structure of a neuronal calcium sensor, guanylate cyclase-activating protein 1.
Structure, 15, 2007
|
|
2ROB
 
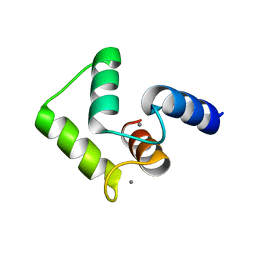 | | Solution structure of calcium bound soybean calmodulin isoform 4 C-terminal domain | | Descriptor: | CALCIUM ION, Calmodulin | | Authors: | Ishida, H, Huang, H, Yamniuk, A.P, Takaya, Y, Vogel, H.J. | | Deposit date: | 2008-03-14 | | Release date: | 2008-04-08 | | Last modified: | 2022-03-16 | | Method: | SOLUTION NMR | | Cite: | The solution structures of two soybean calmodulin isoforms provide a structural basis for their selective target activation properties
J.Biol.Chem., 283, 2008
|
|
2K2F
 
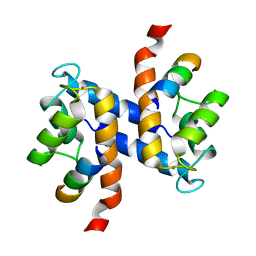 | |
3CR5
 
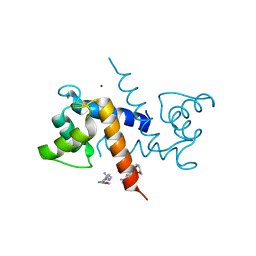 | | X-ray structure of bovine Pnt-Zn(2+),Ca(2+)-S100B | | Descriptor: | 1,5-BIS(4-AMIDINOPHENOXY)PENTANE, CALCIUM ION, Protein S100-B, ... | | Authors: | Charpentier, T.H. | | Deposit date: | 2008-04-04 | | Release date: | 2008-06-24 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Divalent metal ion complexes of S100B in the absence and presence of pentamidine.
J.Mol.Biol., 382, 2008
|
|
3CR2
 
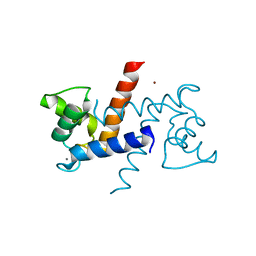 | | X-ray structure of bovine Zn(2+),Ca(2+)-S100B | | Descriptor: | CALCIUM ION, Protein S100-B, ZINC ION | | Authors: | Charpentier, T.H. | | Deposit date: | 2008-04-04 | | Release date: | 2008-06-24 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Divalent metal ion complexes of S100B in the absence and presence of pentamidine.
J.Mol.Biol., 382, 2008
|
|
3CR4
 
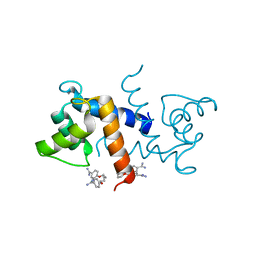 | | X-ray structure of bovine Pnt,Ca(2+)-S100B | | Descriptor: | 1,5-BIS(4-AMIDINOPHENOXY)PENTANE, CALCIUM ION, Protein S100-B | | Authors: | Charpentier, T.H. | | Deposit date: | 2008-04-04 | | Release date: | 2008-08-05 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Divalent metal ion complexes of S100B in the absence and presence of pentamidine.
J.Mol.Biol., 382, 2008
|
|
3CZT
 
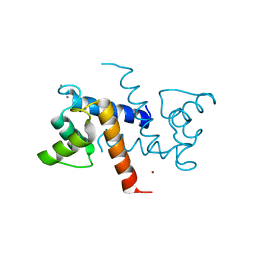 | | Crystal Structure of S100B in the Calcium and Zinc Loaded State at pH 9 | | Descriptor: | CALCIUM ION, Protein S100-B, ZINC ION | | Authors: | Ostendorp, T, Diez, J, Heizmann, C.W, Fritz, G. | | Deposit date: | 2008-04-30 | | Release date: | 2009-04-14 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | The crystal structures of human S100B in the zinc- and calcium-loaded state at three pH values reveal zinc ligand swapping.
Biochim.Biophys.Acta, 1813, 2011
|
|
3D10
 
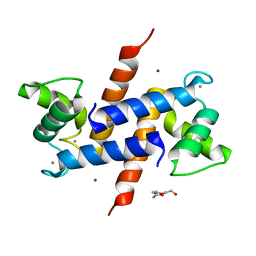 | | Crystal Structure of S100B in the Calcium and Zinc Loaded State at pH 10.0 | | Descriptor: | CALCIUM ION, Protein S100-B, TRIETHYLENE GLYCOL, ... | | Authors: | Ostendorp, T, Diez, J, Heizmann, C.W, Fritz, G. | | Deposit date: | 2008-05-02 | | Release date: | 2009-04-14 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | The crystal structures of human S100B in the zinc- and calcium-loaded state at three pH values reveal zinc ligand swapping.
Biochim.Biophys.Acta, 1813, 2011
|
|
3D0Y
 
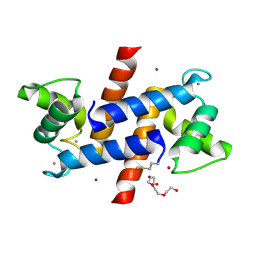 | | Crystal Structure of S100B in the Calcium and Zinc Loaded State at pH 6.5 | | Descriptor: | CALCIUM ION, Protein S100-B, TETRAETHYLENE GLYCOL, ... | | Authors: | Ostendorp, T, Diez, J, Heizmann, C.W, Fritz, G. | | Deposit date: | 2008-05-02 | | Release date: | 2009-04-14 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The crystal structures of human S100B in the zinc- and calcium-loaded state at three pH values reveal zinc ligand swapping.
Biochim.Biophys.Acta, 1813, 2011
|
|
2K7C
 
 | |
2K7D
 
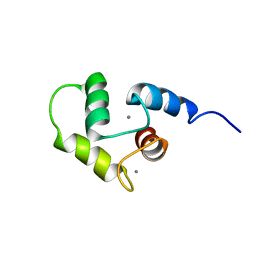 | | NMR Structure of Ca2+-bound CaBP1 C-domain | | Descriptor: | CALCIUM ION, Calcium-binding protein 1 | | Authors: | Ames, J. | | Deposit date: | 2008-08-08 | | Release date: | 2008-11-25 | | Last modified: | 2022-03-16 | | Method: | SOLUTION NMR | | Cite: | NMR Structure of Ca2+-bound CaBP1 C-domain
To be Published
|
|
2K7O
 
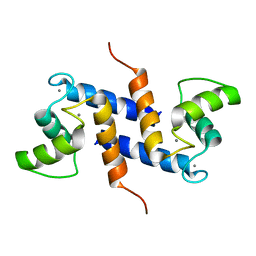 | | Ca2+-S100B, refined with RDCs | | Descriptor: | CALCIUM ION, Protein S100-B | | Authors: | Wright, N.T, Inman, K.G, Levine, J.A, Weber, D.J. | | Deposit date: | 2008-08-17 | | Release date: | 2008-11-18 | | Last modified: | 2022-03-16 | | Method: | SOLUTION NMR | | Cite: | Refinement of the solution structure and dynamic properties of Ca(2+)-bound rat S100B.
J.Biomol.Nmr, 42, 2008
|
|
2W49
 
 | | ISOMETRICALLY CONTRACTING INSECT ASYNCHRONOUS FLIGHT MUSCLE | | Descriptor: | ACTIN, ALPHA SKELETAL MUSCLE, CALCIUM ION, ... | | Authors: | Wu, S, Liu, J, Reedy, M.C, Tregear, R.T, Winkler, H, Franzini-Armstrong, C, Sasaki, H, Lucaveche, C, Goldman, Y.E, Reedy, M.K, Taylor, K.A. | | Deposit date: | 2008-11-24 | | Release date: | 2010-05-05 | | Last modified: | 2019-10-23 | | Method: | ELECTRON MICROSCOPY (35 Å) | | Cite: | Electron Tomography of Cryofixed, Isometrically Contracting Insect Flight Muscle Reveals Novel Actin-Myosin Interactions
Plos One, 5, 2010
|
|
2W4U
 
 | | Isometrically contracting insect asynchronous flight muscle quick frozen after a length step | | Descriptor: | ACTIN, ALPHA SKELETAL MUSCLE, TROPOMYOSIN ALPHA-1 CHAIN, ... | | Authors: | Wu, S, Liu, J, Reedy, M.C, Tregear, R.T, Winkler, H, Franzini-Armstrong, C, Sasaki, H, Lucaveche, C, Goldman, Y.E, Reedy, M.K, Taylor, K.A. | | Deposit date: | 2008-12-02 | | Release date: | 2010-08-25 | | Last modified: | 2019-10-23 | | Method: | ELECTRON MICROSCOPY (35 Å) | | Cite: | Structural Changes in Isometrically Contracting Insect Flight Muscle Trapped Following a Mechanical Perturbation.
Plos One, 7, 2012
|
|
2KBM
 
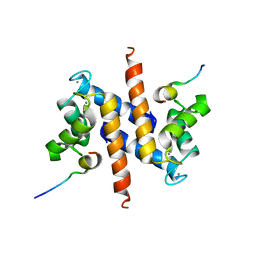 | | Ca-S100A1 interacting with TRTK12 | | Descriptor: | CALCIUM ION, F-actin-capping protein subunit alpha-2, Protein S100-A1 | | Authors: | Wright, N.T, Varney, K.M, Cannon, B.R, Morgan, M, Weber, D.J. | | Deposit date: | 2008-12-02 | | Release date: | 2009-02-10 | | Last modified: | 2018-08-08 | | Method: | SOLUTION NMR | | Cite: | Solution structure of Ca-S100A1-TRTK12
To be Published
|
|
3FWB
 
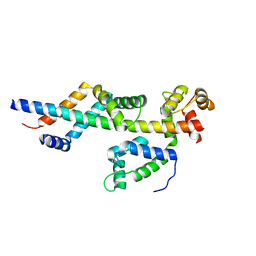 | | Sac3:Sus1:Cdc31 complex | | Descriptor: | Cell division control protein 31, Nuclear mRNA export protein SAC3, Protein SUS1 | | Authors: | Stewart, M, Jani, D. | | Deposit date: | 2009-01-17 | | Release date: | 2009-04-14 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Sus1, Cdc31, and the Sac3 CID region form a conserved interaction platform that promotes nuclear pore association and mRNA export.
Mol.Cell, 33, 2009
|
|
3FWC
 
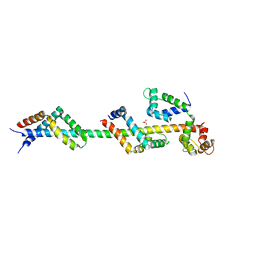 | | Sac3:Sus1:Cdc31 complex | | Descriptor: | Cell division control protein 31, Nuclear mRNA export protein SAC3, Protein SUS1, ... | | Authors: | Stewart, M, Jani, D. | | Deposit date: | 2009-01-17 | | Release date: | 2009-04-14 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Sus1, Cdc31, and the Sac3 CID region form a conserved interaction platform that promotes nuclear pore association and mRNA export.
Mol.Cell, 33, 2009
|
|
3GK2
 
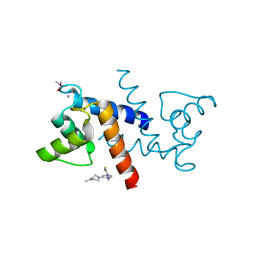 | | X-ray structure of bovine SBi279,Ca(2+)-S100B | | Descriptor: | (Z)-2-[2-(4-methylpiperazin-1-yl)benzyl]diazenecarbothioamide, CACODYLATE ION, CALCIUM ION, ... | | Authors: | Charpentier, T.H, Weber, D.J, Toth, E.A. | | Deposit date: | 2009-03-09 | | Release date: | 2009-06-09 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.984 Å) | | Cite: | Small molecules bound to unique sites in the target protein binding cleft of calcium-bound S100B as characterized by nuclear magnetic resonance and X-ray crystallography.
Biochemistry, 48, 2009
|
|
3GK4
 
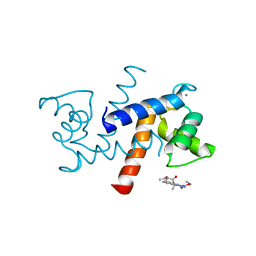 | | X-ray structure of bovine SBi523,Ca(2+)-S100B | | Descriptor: | CALCIUM ION, Protein S100-B, ethyl 5-{[(1R)-1-(ethoxycarbonyl)-2-oxopropyl]sulfanyl}-1,2-dihydro[1,2,3]triazolo[1,5-a]quinazoline-3-carboxylate | | Authors: | Charpentier, T.H, Weber, D.J, Toth, E.A. | | Deposit date: | 2009-03-09 | | Release date: | 2009-06-09 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Small molecules bound to unique sites in the target protein binding cleft of calcium-bound S100B as characterized by nuclear magnetic resonance and X-ray crystallography.
Biochemistry, 48, 2009
|
|
3GK1
 
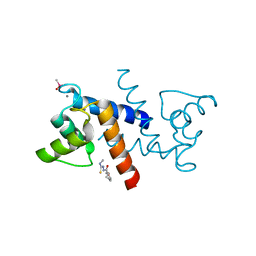 | | X-ray structure of bovine SBi132,Ca(2+)-S100B | | Descriptor: | 2-[(5-hex-1-yn-1-ylfuran-2-yl)carbonyl]-N-methylhydrazinecarbothioamide, CACODYLATE ION, CALCIUM ION, ... | | Authors: | Charpentier, T.H, Weber, D.J, Toth, E.A. | | Deposit date: | 2009-03-09 | | Release date: | 2009-06-09 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Small molecules bound to unique sites in the target protein binding cleft of calcium-bound S100B as characterized by nuclear magnetic resonance and X-ray crystallography.
Biochemistry, 48, 2009
|
|
3HCM
 
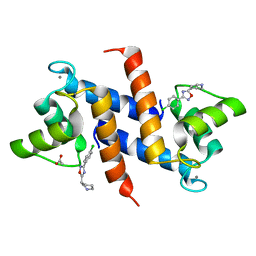 | | Crystal structure of human S100B in complex with S45 | | Descriptor: | (3R)-3-[3-(4-chlorophenyl)-1,2,4-oxadiazol-5-yl]piperidine, ACETATE ION, CALCIUM ION, ... | | Authors: | Mangani, S, Cesari, L. | | Deposit date: | 2009-05-06 | | Release date: | 2010-02-02 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Fragmenting the S100B-p53 Interaction: Combined Virtual/Biophysical Screening Approaches to Identify Ligands
Chemmedchem, 5, 2010
|
|
2KN2
 
 | |
