7SK9
 
 | | Cryo-EM structure of human ACKR3 in complex with a small molecule partial agonist CCX662, and an intracellular Fab | | Descriptor: | (1R)-4-[7-(3-carboxypropoxy)-6-methylquinolin-8-yl]-1-{[2-(4-hydroxypiperidin-1-yl)-1,3-thiazol-4-yl]methyl}-1,4-diazepan-1-ium, Atypical chemokine receptor 3, CHOLESTEROL, ... | | Authors: | Yen, Y.C, Schafer, C.T, Gustavsson, M, Handel, T.M, Tesmer, J.J.G. | | Deposit date: | 2021-10-19 | | Release date: | 2022-07-27 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structures of atypical chemokine receptor 3 reveal the basis for its promiscuity and signaling bias.
Sci Adv, 8, 2022
|
|
7C4S
 
 | | Sphingosine-1-phosphate receptor 3 with a natural ligand. | | Descriptor: | (2S,3R,4E)-2-amino-3-hydroxyoctadec-4-en-1-yl dihydrogen phosphate, Antibody Fab fragment heavy chain, Antibody Fab fragment light chain, ... | | Authors: | Iwata, S, Maeda, S, Luo, F, Nango, E, hirata, K, Asada, H. | | Deposit date: | 2020-05-18 | | Release date: | 2021-06-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Endogenous agonist-bound S1PR3 structure reveals determinants of G protein-subtype bias.
Sci Adv, 7, 2021
|
|
6I9K
 
 | | Crystal structure of Jumping Spider Rhodopsin-1 bound to 9-cis retinal | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, Kumopsin1, RETINAL | | Authors: | Varma, N, Mutt, E, Muehle, J, Panneels, V, Terakita, A, Deupi, X, Nogly, P, Schertler, F.X.G, Lesca, E. | | Deposit date: | 2018-11-23 | | Release date: | 2019-07-03 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.145 Å) | | Cite: | Crystal structure of jumping spider rhodopsin-1 as a light sensitive GPCR.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6IBB
 
 | | Crystal structure of the rat isoform of the succinate receptor SUCNR1 (GPR91) in complex with a nanobody | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, (2~{S},5~{R})-hexane-2,5-diol, CHOLESTEROL, ... | | Authors: | Haffke, M, Jaakola, V.-P. | | Deposit date: | 2018-11-29 | | Release date: | 2019-08-14 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Structural basis of species-selective antagonist binding to the succinate receptor.
Nature, 574, 2019
|
|
6IGL
 
 | | Crystal Structure of human ETB receptor in complex with IRL1620 | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, CITRIC ACID, Endothelin receptor type B,Endolysin,Endothelin receptor type B, ... | | Authors: | Shihoya, W, Izume, T, Inoue, A, Yamashita, K, kadji, F.M.N, Hirata, K, Aoki, J, Nishizawa, T, Nureki, O. | | Deposit date: | 2018-09-25 | | Release date: | 2018-11-21 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structures of human ETBreceptor provide mechanistic insight into receptor activation and partial activation.
Nat Commun, 9, 2018
|
|
6KR8
 
 | |
6KUY
 
 | | Crystal structure of the alpha2A adrenergic receptor in complex with a partial agonist | | Descriptor: | (2~{S})-4-fluoranyl-2-(1~{H}-imidazol-5-yl)-1-propan-2-yl-2,3-dihydroindole, Alpha2A adrenergic receptor, DI(HYDROXYETHYL)ETHER | | Authors: | Qu, L, Zhou, Q.T, Wu, D, Zhao, S.W. | | Deposit date: | 2019-09-02 | | Release date: | 2019-12-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Crystal structures of the alpha2A adrenergic receptor in complex with a partial agonist
To Be Published
|
|
6KUX
 
 | | Crystal structures of the alpha2A adrenergic receptor in complex with an antagonist RSC. | | Descriptor: | (8~{a}~{R},12~{a}~{S},13~{a}~{S})-12-ethylsulfonyl-3-methoxy-5,6,8,8~{a},9,10,11,12~{a},13,13~{a}-decahydroisoquinolino[2,1-g][1,6]naphthyridine, CITRATE ANION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Qu, L, Zhou, Q.T, Wu, D, Zhao, S.W. | | Deposit date: | 2019-09-02 | | Release date: | 2019-12-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structures of the alpha2A adrenergic receptor in complex with an antagonist RSC.
To Be Published
|
|
6N48
 
 | | Structure of beta2 adrenergic receptor bound to BI167107, Nanobody 6B9, and a positive allosteric modulator | | Descriptor: | (2S)-2,3-dihydroxypropyl (7Z)-tetradec-7-enoate, 8-[(1R)-2-{[1,1-dimethyl-2-(2-methylphenyl)ethyl]amino}-1-hydroxyethyl]-5-hydroxy-2H-1,4-benzoxazin-3(4H)-one, Camelid Antibody Fragment, ... | | Authors: | Liu, X, Masoudi, A, Kahsai, A.W, Huang, L.Y, Pani, B, Hirata, K, Ahn, S, Lefkowitz, R.J, Kobilka, B.K. | | Deposit date: | 2018-11-17 | | Release date: | 2019-06-26 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Mechanism of beta2AR regulation by an intracellular positive allosteric modulator.
Science, 364, 2019
|
|
8XBI
 
 | | Human GPR34 -Gi complex bound to M1, receptor focused | | Descriptor: | (2~{S})-2-azanyl-3-[[(2~{R})-1-ethoxy-3-[3-[2-[(3-phenoxyphenyl)methoxy]phenyl]propanoyloxy]propan-2-yl]oxy-oxidanyl-phosphoryl]oxy-propanoic acid, Probable G-protein coupled receptor 34 | | Authors: | Kawahara, R, Shihoya, W, Nureki, O. | | Deposit date: | 2023-12-06 | | Release date: | 2023-12-27 | | Method: | ELECTRON MICROSCOPY (3.06 Å) | | Cite: | Cryo-EM structure of the Human GPR34-Gi complex bound to S3E-LysoPS
To Be Published
|
|
8WCC
 
 | | Cryo-EM structure of the CHA-bound mTAAR1 complex | | Descriptor: | CYCLOHEXYLAMMONIUM ION, Trace amine-associated receptor 1 | | Authors: | Rong, N.K, Guo, L.L, Zhang, M.H, Li, Q, Yang, F, Sun, J.P. | | Deposit date: | 2023-09-11 | | Release date: | 2023-12-27 | | Method: | ELECTRON MICROSCOPY (3.04 Å) | | Cite: | Structural and signaling mechanisms of TAAR1 enabled preferential agonist design.
Cell, 186, 2023
|
|
8W8S
 
 | | Cryo-EM structure of the AA14-bound GPR101 complex | | Descriptor: | 1-(4-methylpyridin-2-yl)-3-[3-(trifluoromethyl)phenyl]thiourea, Probable G-protein coupled receptor 101 | | Authors: | Sun, J.P, Yu, X, Gao, N, Yang, F, Wang, J.Y, Yang, Z, Guan, Y, Wang, G.P. | | Deposit date: | 2023-09-04 | | Release date: | 2024-01-03 | | Last modified: | 2024-04-10 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structure of GPR101-Gs enables identification of ligands with rejuvenating potential.
Nat.Chem.Biol., 20, 2024
|
|
8WDT
 
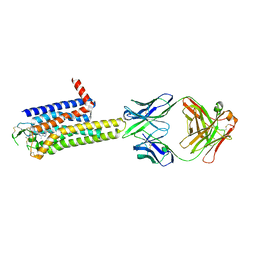 | | Crystal structure of the human adenosine A2A receptor in complex with photoresponsive ligand photoNECA(blue) | | Descriptor: | (2S,3S,4R,5R)-5-(6-amino-2-((E)-phenyldiazenyl)-9H-purin-9-yl)-N-ethyl-3,4-dihydroxytetrahydrofuran-2-carboxamide, Adenosine receptor A2a, Antibody Fab fragment heavy chain, ... | | Authors: | Araya, T, Asada, H, Iwata, S, Im, D.H. | | Deposit date: | 2023-09-16 | | Release date: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.34 Å) | | Cite: | Crystal structure reveals the binding mode and selectivity of a photoswitchable ligand for the adenosine A 2A receptor.
Biochem.Biophys.Res.Commun., 695, 2023
|
|
7SRQ
 
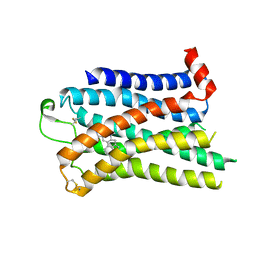 | | 5-HT2B receptor bound to LSD obtained by cryo-electron microscopy (cryoEM) | | Descriptor: | (8alpha)-N,N-diethyl-6-methyl-9,10-didehydroergoline-8-carboxamide, 2-acetamido-2-deoxy-beta-D-glucopyranose, 5-hydroxytryptamine receptor 2B | | Authors: | Barros-Alvarez, X, Cao, C, Panova, O, Roth, B.L, Skiniotis, G. | | Deposit date: | 2021-11-08 | | Release date: | 2022-09-21 | | Last modified: | 2022-10-19 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Signaling snapshots of a serotonin receptor activated by the prototypical psychedelic LSD.
Neuron, 110, 2022
|
|
7TUY
 
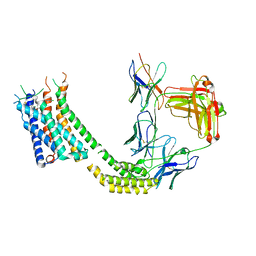 | | Cryo-EM structure of GSK682753A-bound EBI2/GPR183 | | Descriptor: | 8-[(2E)-3-(4-chlorophenyl)prop-2-enoyl]-3-[(3,4-dichlorophenyl)methyl]-1-oxa-3,8-diazaspiro[4.5]decan-2-one, G-protein coupled receptor 183,Soluble cytochrome b562 fusion, anti-BRIL Fab Heavy chain, ... | | Authors: | Chen, H, Huang, W, Li, X. | | Deposit date: | 2022-02-03 | | Release date: | 2022-04-13 | | Last modified: | 2022-07-20 | | Method: | ELECTRON MICROSCOPY (2.98 Å) | | Cite: | Structures of oxysterol sensor EBI2/GPR183, a key regulator of the immune response.
Structure, 30, 2022
|
|
7UL5
 
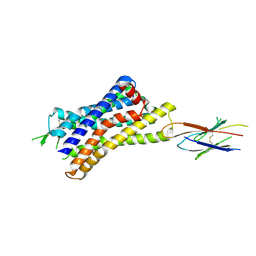 | |
7UL2
 
 | | CryoEM Structure of Inactive NTSR1 Bound to SR48692 and Nb6 | | Descriptor: | 2-[[1-(7-chloranylquinolin-4-yl)-5-(2,6-dimethoxyphenyl)pyrazol-3-yl]carbonylamino]adamantane-2-carboxylic acid, Nanobody 6, Neurotensin receptor 1, ... | | Authors: | Robertson, M.J, Skiniotis, G. | | Deposit date: | 2022-04-03 | | Release date: | 2022-06-29 | | Last modified: | 2022-12-28 | | Method: | ELECTRON MICROSCOPY (2.4 Å) | | Cite: | Structure determination of inactive-state GPCRs with a universal nanobody.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7UL3
 
 | | CryoEM Structure of Inactive H2R Bound to Famotidine, Nb6M, and NabFab | | Descriptor: | Histamine H2 receptor, NabFab HC, NabFab LC, ... | | Authors: | Robertson, M.J, Skiniotis, G. | | Deposit date: | 2022-04-03 | | Release date: | 2022-06-29 | | Last modified: | 2022-12-28 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structure determination of inactive-state GPCRs with a universal nanobody.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7UL4
 
 | | CryoEM Structure of Inactive MOR Bound to Alvimopan and Mb6 | | Descriptor: | Megabody 6, Mu-type opioid receptor, N-[(2S)-2-{[(3R,4R)-4-(3-hydroxyphenyl)-3,4-dimethylpiperidin-1-yl]methyl}-3-phenylpropanoyl]glycine | | Authors: | Robertson, M.J, Skiniotis, G. | | Deposit date: | 2022-04-03 | | Release date: | 2022-06-29 | | Last modified: | 2022-12-28 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structure determination of inactive-state GPCRs with a universal nanobody.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7VV0
 
 | |
7VV6
 
 | | Cryo-EM structure of pseudoallergen receptor MRGPRX2 complex with C48/80 (local) | | Descriptor: | 2-[4-methoxy-3-[[2-methoxy-3-[[2-methoxy-5-[2-(methylamino)ethyl]phenyl]methyl]-5-[2-(methylamino)ethyl]phenyl]methyl]phenyl]-~{N}-methyl-ethanamine, CHOLESTEROL, Mas-related G-protein coupled receptor member X2 | | Authors: | Li, Y, Yang, F. | | Deposit date: | 2021-11-04 | | Release date: | 2021-12-01 | | Last modified: | 2022-07-20 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structure, function and pharmacology of human itch receptor complexes.
Nature, 600, 2021
|
|
8HN1
 
 | |
7WC5
 
 | | Crystal structure of serotonin 2A receptor in complex with psilocin | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 3-[2-(dimethylamino)ethyl]-1~{H}-indol-4-ol, 5-hydroxytryptamine receptor 2A, ... | | Authors: | Cao, D, Yu, J, Wang, H, Luo, Z, Liu, X, He, L, Qi, J, Fan, L, Tang, L, Chen, Z, Li, J, Cheng, J, Wang, S. | | Deposit date: | 2021-12-18 | | Release date: | 2022-01-26 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structure-based discovery of nonhallucinogenic psychedelic analogs.
Science, 375, 2022
|
|
7WC4
 
 | | Crystal structure of serotonin 2A receptor in complex with serotonin | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 5-hydroxytryptamine receptor 2A,5-hydroxytryptamine receptor 2A,5-hydroxytryptamine receptor 2A,Soluble cytochrome b562, CHOLESTEROL, ... | | Authors: | Cao, D, Yu, J, Wang, H, Luo, Z, Liu, X, He, L, Qi, J, Fan, L, Tang, L, Chen, Z, Li, J, Cheng, J, Wang, S. | | Deposit date: | 2021-12-18 | | Release date: | 2022-01-26 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structure-based discovery of nonhallucinogenic psychedelic analogs.
Science, 375, 2022
|
|
8IWM
 
 | | Cryo-EM structure of the PEA-bound mTAAR9 complex | | Descriptor: | 2-PHENYLETHYLAMINE, Trace amine-associated receptor 9 | | Authors: | Sun, J.P, Li, Q, Yang, F, Xu, Y.F, Guo, L.L, Lian, S, Zhang, M.H, Rong, N.K. | | Deposit date: | 2023-03-30 | | Release date: | 2023-05-31 | | Last modified: | 2023-06-14 | | Method: | ELECTRON MICROSCOPY (3.17 Å) | | Cite: | Structural basis of amine odorant perception by a mammal olfactory receptor.
Nature, 618, 2023
|
|
