6PMH
 
 | | Structure of Epimerase Mth375 from the thermophilic pseudomurein-containing methanogen Methanothermobacter thermautotrophicus | | Descriptor: | ADENOSINE MONOPHOSPHATE, CHLORIDE ION, GLYCEROL, ... | | Authors: | Carbone, V, Schofield, L.R, Sutherland-Smith, A.J, Ronimus, R.S. | | Deposit date: | 2019-07-01 | | Release date: | 2020-07-08 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of a UDP-GALE 4-epimerase (Mth375) from the thermophilic pseudomurein-containing methanogen Methanothermobacter thermautotrophicus
To Be Published
|
|
4TWR
 
 | | Structure of UDP-glucose 4-epimerase from Brucella abortus | | Descriptor: | NAD binding site:NAD-dependent epimerase/dehydratase:UDP-glucose 4-epimerase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ZINC ION | | Authors: | Horanyi, P.S, Abendroth, J, Lorimer, D, Edwards, T, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2014-07-01 | | Release date: | 2014-10-08 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of UDP-glucose 4-epimerase from Brucella melitensis
To Be Published
|
|
6WJ9
 
 | | UDP-GlcNAc C4-epimerase mutant S121A/Y146F from Pseudomonas protegens in complex with UDP-GlcNAc | | Descriptor: | NAD-dependent epimerase/dehydratase family protein, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, URIDINE-DIPHOSPHATE-N-ACETYLGLUCOSAMINE | | Authors: | Marmont, L.S, Willams, R.J, Whitney, J.C, Whitfield, G.B, Robinson, H, Parsek, M.R, Nitz, M, Harrison, J.J, Howell, P.L. | | Deposit date: | 2020-04-13 | | Release date: | 2020-07-08 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | PelX is a UDP-N-acetylglucosamine C4-epimerase involved in Pel polysaccharide-dependent biofilm formation.
J.Biol.Chem., 295, 2020
|
|
6JYG
 
 | | Crystal Structure of L-threonine dehydrogenase from Phytophthora infestans | | Descriptor: | 3,6,9,12,15,18,21-HEPTAOXATRICOSANE-1,23-DIOL, CITRATE ANION, L-threonine 3-dehydrogenase, ... | | Authors: | Yoneda, K, Sakuraba, H, Ohshima, T. | | Deposit date: | 2019-04-26 | | Release date: | 2020-04-01 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Catalytic properties and crystal structure of UDP-galactose 4-epimerase-like l-threonine 3-dehydrogenase from Phytophthora infestans.
Enzyme.Microb.Technol., 140, 2020
|
|
6WJB
 
 | | UDP-GlcNAc C4-epimerase from Pseudomonas protegens in complex with NAD and UDP-GlcNAc | | Descriptor: | NAD-dependent epimerase/dehydratase family protein, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, URIDINE-DIPHOSPHATE-N-ACETYLGLUCOSAMINE | | Authors: | Marmont, L.S, Pfoh, R, Robinson, H, Howell, P.L. | | Deposit date: | 2020-04-13 | | Release date: | 2020-07-08 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | PelX is a UDP-N-acetylglucosamine C4-epimerase involved in Pel polysaccharide-dependent biofilm formation.
J.Biol.Chem., 295, 2020
|
|
6KWS
 
 | |
6KV9
 
 | | MoeE5 in complex with UDP-glucuronic acid and NAD | | Descriptor: | MoeE5, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, URIDINE-5'-DIPHOSPHATE-GLUCURONIC ACID | | Authors: | Ko, T.-P, Liu, W, Sun, H, Liu, W, Chen, C.-C, Guo, R.-T. | | Deposit date: | 2019-09-03 | | Release date: | 2019-11-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Structure of an antibiotic-synthesizing UDP-glucuronate 4-epimerase MoeE5 in complex with substrate.
Biochem.Biophys.Res.Commun., 521, 2020
|
|
6KWT
 
 | |
6KVC
 
 | | MoeE5 in complex with UDP-glucose and NAD | | Descriptor: | MoeE5, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, URIDINE-5'-DIPHOSPHATE-GLUCOSE | | Authors: | Ko, T.-P, Liu, W, Sun, H, Liu, W, Chen, C.-C, Guo, R.-T. | | Deposit date: | 2019-09-04 | | Release date: | 2019-11-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Structure of an antibiotic-synthesizing UDP-glucuronate 4-epimerase MoeE5 in complex with substrate.
Biochem.Biophys.Res.Commun., 521, 2020
|
|
6MFH
 
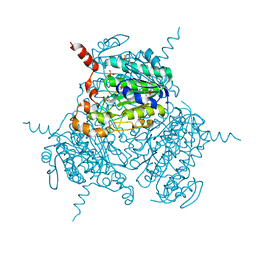 | | Mutated Uronate Dehydrogenase | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, Mutated Uronate Dehydrogenase, PHOSPHATE ION | | Authors: | Sankaran, B, Pereira, J.H, Chan, V, Zwart, P.H, Wagschal, K. | | Deposit date: | 2018-09-11 | | Release date: | 2019-09-11 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Chromohalobacter salixigens uronate dehydrogenase: Directed evolution for improved thermal stability and mutant CsUDH-inc X-ray crystal structure
Process Biochem, 2020
|
|
5K50
 
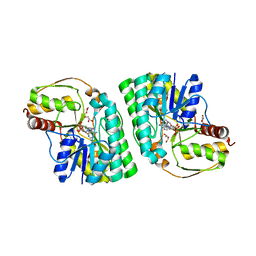 | | Three-dimensional structure of L-threonine 3-dehydrogenase from Trypanosoma brucei bound to NAD+ and L-allo-threonine refined to 2.23 angstroms | | Descriptor: | ACETATE ION, ALLO-THREONINE, GLYCEROL, ... | | Authors: | Adjogatse, E.A, Erskine, P.T, Cooper, J.B. | | Deposit date: | 2016-05-22 | | Release date: | 2017-11-15 | | Last modified: | 2018-10-10 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Structure and function of L-threonine-3-dehydrogenase from the parasitic protozoan Trypanosoma brucei revealed by X-ray crystallography and geometric simulations.
Acta Crystallogr D Struct Biol, 74, 2018
|
|
5K4Y
 
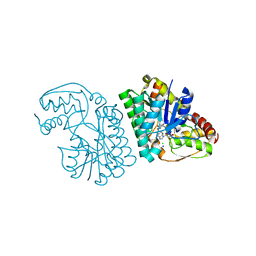 | | Three-dimensional structure of L-threonine 3-dehydrogenase from Trypanosoma brucei refined to 1.77 angstroms | | Descriptor: | ACETATE ION, CHLORIDE ION, GLYCEROL, ... | | Authors: | Adjogatse, E.A, Erskine, P.T, Cooper, J.B. | | Deposit date: | 2016-05-22 | | Release date: | 2018-01-17 | | Last modified: | 2018-10-10 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Structure and function of L-threonine-3-dehydrogenase from the parasitic protozoan Trypanosoma brucei revealed by X-ray crystallography and geometric simulations.
Acta Crystallogr D Struct Biol, 74, 2018
|
|
6D2V
 
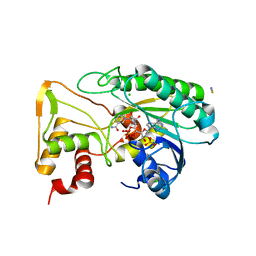 | | Apo Structure of TerB, an NADP Dependent Oxidoreductase in the Terfestatin Biosynthesis Pathway | | Descriptor: | CHLORIDE ION, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, THIOCYANATE ION, ... | | Authors: | Clinger, J.A, Elshahawi, S.I, Zhang, Y, Hall, R.P, Liu, Y, Thorson, J.S, Phillips Jr, G.N. | | Deposit date: | 2018-04-14 | | Release date: | 2018-06-13 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.901 Å) | | Cite: | Structure and Function of Terfestatin Biosynthesis Enzymes TerB and TerC
To Be Published
|
|
6DNT
 
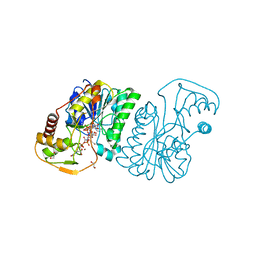 | | UDP-N-acetylglucosamine 4-epimerase from Methanobrevibacter ruminantium M1 in complex with UDP-N-acetylmuramic acid | | Descriptor: | (2R)-2-{[(2R,3R,4R,5S,6R)-3-(acetylamino)-2-{[(S)-{[(R)-{[(2R,3S,4R,5R)-5-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)-3,4-dihydroxytetrahydrofuran-2-yl]methoxy}(hydroxy)phosphoryl]oxy}(hydroxy)phosphoryl]oxy}-5-hydroxy-6-(hydroxymethyl)tetrahydro-2H-pyran-4-yl]oxy}propanoic acid, 1,2-ETHANEDIOL, NAD-dependent epimerase/dehydratase, ... | | Authors: | Carbone, V, Schofield, L.R, Sang, C, Sutherland-Smith, A.J, Ronimus, R.S. | | Deposit date: | 2018-06-07 | | Release date: | 2018-10-03 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Structural determination of archaeal UDP-N-acetylglucosamine 4-epimerase from Methanobrevibacter ruminantium M1 in complex with the bacterial cell wall intermediate UDP-N-acetylmuramic acid.
Proteins, 86, 2018
|
|
1UDA
 
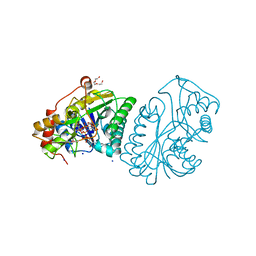 | |
5L9A
 
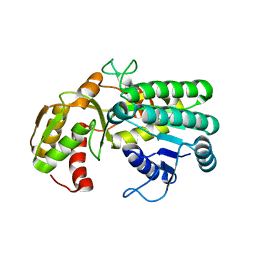 | | L-threonine dehydrogenase from trypanosoma brucei. | | Descriptor: | ACETATE ION, L-threonine 3-dehydrogenase | | Authors: | Erskine, P.T, Cooper, J.B, Adjogatse, E, Kelly, J, Wood, S.P. | | Deposit date: | 2016-06-09 | | Release date: | 2016-06-22 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structure and function of L-threonine-3-dehydrogenase from the parasitic protozoan Trypanosoma brucei revealed by X-ray crystallography and geometric simulations.
Acta Crystallogr D Struct Biol, 74, 2018
|
|
5LC1
 
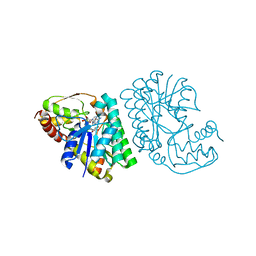 | | L-threonine dehydrogenase from Trypanosoma brucei with NAD and the inhibitor pyruvate bound. | | Descriptor: | ACETATE ION, BETA-MERCAPTOETHANOL, L-threonine 3-dehydrogenase, ... | | Authors: | Erskine, P.T, Adjogatse, E, Wood, S.P, Cooper, J.B. | | Deposit date: | 2016-06-18 | | Release date: | 2016-07-27 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure and function of L-threonine-3-dehydrogenase from the parasitic protozoan Trypanosoma brucei revealed by X-ray crystallography and geometric simulations.
Acta Crystallogr D Struct Biol, 74, 2018
|
|
4YRB
 
 | | mouse TDH mutant R180K with NAD+ bound | | Descriptor: | L-threonine 3-dehydrogenase, mitochondrial, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | He, C, Li, F. | | Deposit date: | 2015-03-14 | | Release date: | 2016-02-03 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | Structural insights on mouse l-threonine dehydrogenase: A regulatory role of Arg180 in catalysis
J.Struct.Biol., 192, 2015
|
|
4YRD
 
 | | Crystal structure of CapF with inhibitor 3-isopropenyl-tropolone | | Descriptor: | 2-hydroxy-3-(prop-1-en-2-yl)cyclohepta-2,4,6-trien-1-one, Capsular polysaccharide synthesis enzyme Cap5F, ZINC ION | | Authors: | Nakano, K, Chigira, T, Miyafusa, T, Nagatoishi, S, Caaveiro, J.M.M, Tsumoto, K. | | Deposit date: | 2015-03-15 | | Release date: | 2015-10-21 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | Discovery and characterization of natural tropolones as inhibitors of the antibacterial target CapF from Staphylococcus aureus.
Sci Rep, 5, 2015
|
|
1U9J
 
 | |
6EL3
 
 | | Structure of Progesterone 5beta-Reductase from Arabidopsis thaliana in complex with NADP | | Descriptor: | 3-oxo-Delta(4,5)-steroid 5-beta-reductase, CHLORIDE ION, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Muller, Y.A, Schmidt, K, Egerer-Sieber, C. | | Deposit date: | 2017-09-27 | | Release date: | 2018-09-05 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.899 Å) | | Cite: | PRISEs (progesterone 5 beta-reductase and/or iridoid synthase-like 1,4-enone reductases): Catalytic and substrate promiscuity allows for realization of multiple pathways in plant metabolism.
Phytochemistry, 156, 2018
|
|
1UDB
 
 | |
1E7Q
 
 | | GDP 4-keto-6-deoxy-D-mannose epimerase reductase S107A | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ACETYLPHOSPHATE, GDP-FUCOSE SYNTHETASE, ... | | Authors: | Rosano, C, Izzo, G, Bolognesi, M. | | Deposit date: | 2000-09-07 | | Release date: | 2000-10-18 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Probing the Catalytic Mechanism of Gdp-4-Keto-6-Deoxy-D-Mannose Epimerase/Reductase by Kinetic and Crystallographic Characterization of Site-Specific Mutants
J.Mol.Biol., 303, 2000
|
|
1UDC
 
 | |
1UJM
 
 | |
