8G7M
 
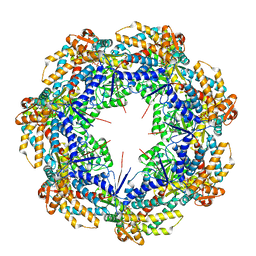 | | ATP-bound mtHsp60 V72I focus | | Descriptor: | 60 kDa heat shock protein, mitochondrial, ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Braxton, J.R, Shao, H, Tse, E, Gestwicki, J.E, Southworth, D.R. | | Deposit date: | 2023-02-16 | | Release date: | 2023-07-12 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Asymmetric apical domain states of mitochondrial Hsp60 coordinate substrate engagement and chaperonin assembly.
Biorxiv, 2023
|
|
8G7L
 
 | | ATP-bound mtHsp60 V72I | | Descriptor: | 60 kDa heat shock protein, mitochondrial, ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Braxton, J.R, Shao, H, Tse, E, Gestwicki, J.E, Southworth, D.R. | | Deposit date: | 2023-02-16 | | Release date: | 2023-07-12 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Asymmetric apical domain states of mitochondrial Hsp60 coordinate substrate engagement and chaperonin assembly.
Biorxiv, 2023
|
|
7NVL
 
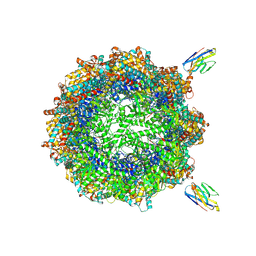 | | Human TRiC complex in closed state with nanobody bound (Consensus Map) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ALUMINUM FLUORIDE, MAGNESIUM ION, ... | | Authors: | Kelly, J.J, Chi, G, Bulawa, C, Paavilainen, V.O, Bountra, C, Huiskonen, J.T, Yue, W, Structural Genomics Consortium (SGC) | | Deposit date: | 2021-03-15 | | Release date: | 2022-03-02 | | Last modified: | 2022-06-01 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Snapshots of actin and tubulin folding inside the TRiC chaperonin.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7NVO
 
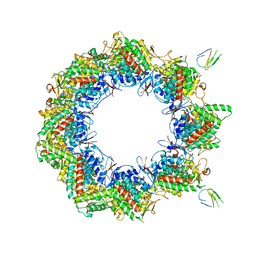 | | Human TRiC complex in open state with nanobody bound | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ALUMINUM FLUORIDE, MAGNESIUM ION, ... | | Authors: | Kelly, J.J, Chi, G, Bulawa, C, Paavilainen, V.O, Bountra, C, Huiskonen, J.T, Yue, W. | | Deposit date: | 2021-03-15 | | Release date: | 2022-03-02 | | Last modified: | 2022-06-01 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Snapshots of actin and tubulin folding inside the TRiC chaperonin.
Nat.Struct.Mol.Biol., 29, 2022
|
|
5UYZ
 
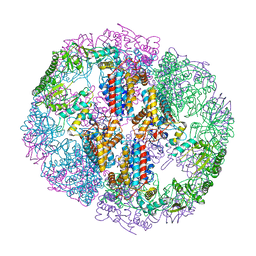 | | Structure of Human T-complex protein 1 subunit epsilon (CCT5) mutant His147Arg | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, T-complex protein 1 subunit epsilon | | Authors: | Pereira, J.H, McAndrew, R.P, Sergeeva, O.A, Ralston, C.Y, King, J.A, Adams, P.D. | | Deposit date: | 2017-02-24 | | Release date: | 2017-07-05 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Structure of the human TRiC/CCT Subunit 5 associated with hereditary sensory neuropathy.
Sci Rep, 7, 2017
|
|
5UYX
 
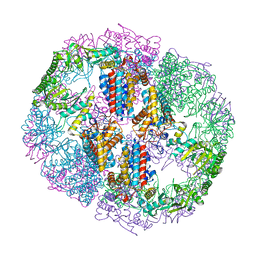 | | Structure of Human T-complex protein 1 subunit epsilon (CCT5) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, T-complex protein 1 subunit epsilon | | Authors: | Pereira, J.H, McAndrew, R.P, Sergeeva, O.A, Ralston, C.Y, King, J.A, Adams, P.D. | | Deposit date: | 2017-02-24 | | Release date: | 2017-07-05 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structure of the human TRiC/CCT Subunit 5 associated with hereditary sensory neuropathy.
Sci Rep, 7, 2017
|
|
5W0S
 
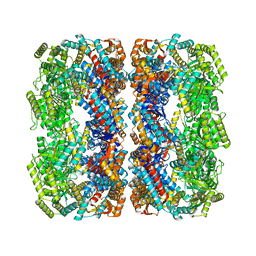 | | GroEL using cryoEM | | Descriptor: | 60 kDa chaperonin | | Authors: | Roh, S.H, Chiu, W. | | Deposit date: | 2017-05-31 | | Release date: | 2017-08-09 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Subunit conformational variation within individual GroEL oligomers resolved by Cryo-EM.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5W74
 
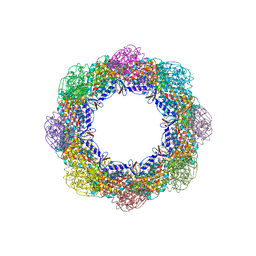 | | Crystal Structure of the Group II Chaperonin from Methanococcus Maripaludis D386ADeltaLid Mutant in the Open, ADP-Bound State | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Chaperonin, MAGNESIUM ION | | Authors: | Dalton, K.M, Lopez, T, Liu, C, Ralston, C.Y, Pereira, J.H, Chartron, J.W, McAndrew, R.P, Douglas, N.R, Adams, P.D, Pande, V.S, Frydman, J. | | Deposit date: | 2017-06-19 | | Release date: | 2018-06-20 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3.65 Å) | | Cite: | The Conformational Cycle of the Group II Chaperonin Termini
To Be Published
|
|
5W79
 
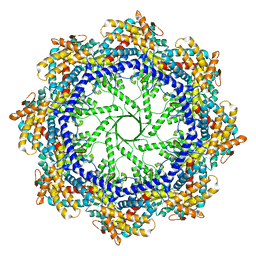 | | Crystal Structure of the Group II Chaperonin from Methanococcus Maripaludis, Cysteine-less mutant in the Apo State | | Descriptor: | Chaperonin, SULFATE ION | | Authors: | Dalton, K.M, Lopez, T, Liu, C, Ralston, C.Y, Pereira, J.H, Chartron, J.W, McAndrew, R.P, Douglas, N.R, Adams, P.D, Pande, V.S, Frydman, J. | | Deposit date: | 2017-06-19 | | Release date: | 2018-06-20 | | Last modified: | 2018-08-29 | | Method: | X-RAY DIFFRACTION (3.122 Å) | | Cite: | The Conformational Cycle of the Group II Chaperonin Termini
To Be Published
|
|
5X9U
 
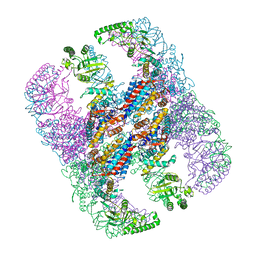 | | Crystal structure of group III chaperonin in the open state | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Thermosome, alpha subunit | | Authors: | An, Y.J, Cha, S.S. | | Deposit date: | 2017-03-09 | | Release date: | 2017-10-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (4.001 Å) | | Cite: | Structural and mechanistic characterization of an archaeal-like chaperonin from a thermophilic bacterium
Nat Commun, 8, 2017
|
|
1XCK
 
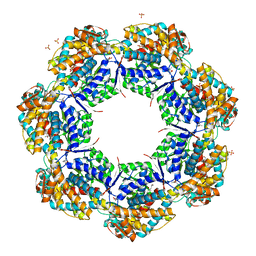 | | Crystal structure of apo GroEL | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 60 kDa chaperonin, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Bartolucci, C, Lamba, D, Grazulis, S, Manakova, E, Heumann, H. | | Deposit date: | 2004-09-02 | | Release date: | 2005-10-25 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.92 Å) | | Cite: | Crystal structure of wild-type chaperonin GroEL
J.Mol.Biol., 354, 2005
|
|
1GRL
 
 | | THE CRYSTAL STRUCTURE OF THE BACTERIAL CHAPERONIN GROEL AT 2.8 ANGSTROMS | | Descriptor: | GROEL (HSP60 CLASS) | | Authors: | Braig, K, Otwinowski, Z, Hegde, R, Boisvert, D.C, Joachimiak, A, Horwich, A.L, Sigler, P.B. | | Deposit date: | 1995-03-07 | | Release date: | 1995-10-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The crystal structure of the bacterial chaperonin GroEL at 2.8 A.
Nature, 371, 1994
|
|
6NRA
 
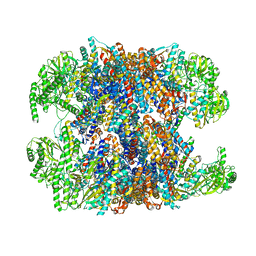 | | hTRiC-hPFD Class1 (No PFD) | | Descriptor: | T-complex protein 1 subunit alpha, T-complex protein 1 subunit beta, T-complex protein 1 subunit delta, ... | | Authors: | Gestaut, D.R, Roh, S.H, Ma, B, Pintilie, G, Joachimiak, L.A, Leitner, A, Walzthoeni, T, Aebersold, R, Chiu, W, Frydman, J. | | Deposit date: | 2019-01-23 | | Release date: | 2019-06-19 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (7.7 Å) | | Cite: | The Chaperonin TRiC/CCT Associates with Prefoldin through a Conserved Electrostatic Interface Essential for Cellular Proteostasis.
Cell, 177, 2019
|
|
1IOK
 
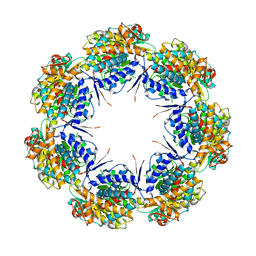 | | CRYSTAL STRUCTURE OF CHAPERONIN-60 FROM PARACOCCUS DENITRIFICANS | | Descriptor: | CHAPERONIN 60 | | Authors: | Fukami, T.A, Yohda, M, Taguchi, H, Yoshida, M, Miki, K. | | Deposit date: | 2001-03-16 | | Release date: | 2001-10-03 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Crystal structure of chaperonin-60 from Paracoccus denitrificans.
J.Mol.Biol., 312, 2001
|
|
1E0R
 
 | | Beta-apical domain of thermosome | | Descriptor: | THERMOSOME | | Authors: | Bosch, G, Baumeister, W, Essen, L.-O. | | Deposit date: | 2000-04-06 | | Release date: | 2000-08-19 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal Structure of the Beta-Apical Domain from Thermosome Reveals Structural Plasticity in Protrusion Region
J.Mol.Biol., 301, 2000
|
|
7V9R
 
 | | Crystal Structure of the heptameric EcHsp60 | | Descriptor: | 60 kDa chaperonin | | Authors: | Lai, M.C, Lin, S.M. | | Deposit date: | 2021-08-26 | | Release date: | 2022-08-31 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Crystal structures of dimeric and heptameric mtHsp60 reveal the mechanism of chaperonin inactivation.
Life Sci Alliance, 6, 2023
|
|
7V98
 
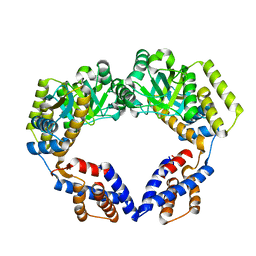 | | Crystal Structure of the Dimeric EcHsp60 | | Descriptor: | 60 kDa chaperonin | | Authors: | Lai, M.C, Lin, S.M. | | Deposit date: | 2021-08-24 | | Release date: | 2022-08-31 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Crystal structures of dimeric and heptameric mtHsp60 reveal the mechanism of chaperonin inactivation.
Life Sci Alliance, 6, 2023
|
|
6QB8
 
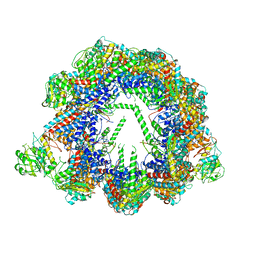 | | Human CCT:mLST8 complex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, T-complex protein 1 subunit alpha, T-complex protein 1 subunit beta, ... | | Authors: | Cuellar, J, Santiago, C, Ludlam, W.G, Bueno-Carrasco, M.T, Valpuesta, J.M, Willardson, B.M. | | Deposit date: | 2018-12-20 | | Release date: | 2019-07-03 | | Last modified: | 2022-03-30 | | Method: | ELECTRON MICROSCOPY (3.97 Å) | | Cite: | Structural and functional analysis of the role of the chaperonin CCT in mTOR complex assembly.
Nat Commun, 10, 2019
|
|
1JON
 
 | |
1KID
 
 | |
1KP8
 
 | |
1FY9
 
 | |
1FYA
 
 | |
1GN1
 
 | | crystal structure of the mouse CCT gamma apical domain (monoclinic) | | Descriptor: | CALCIUM ION, CCT-GAMMA | | Authors: | Pappenberger, G, Wilsher, J.A, Roe, S.M, Willison, K.R, Pearl, L.H. | | Deposit date: | 2001-10-01 | | Release date: | 2002-06-18 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal Structure of the Cct Gamma Apical Domain:: Implications for Substrate Binding to the Eukaryotic Cytosolic Chaperonin
J.Mol.Biol., 318, 2002
|
|
1GML
 
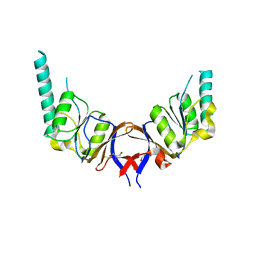 | | crystal structure of the mouse CCT gamma apical domain (triclinic) | | Descriptor: | GLYCEROL, T-COMPLEX PROTEIN 1 SUBUNIT GAMMA | | Authors: | Pappenberger, G, Wilsher, J.A, Roe, S.M, Willison, K.R, Pearl, L.H. | | Deposit date: | 2001-09-17 | | Release date: | 2002-06-18 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structure of the Cct Gamma Apical Domain:: Implications for Substrate Binding to the Eukaryotic Cytosolic Chaperonin
J.Mol.Biol., 318, 2002
|
|
