6QTV
 
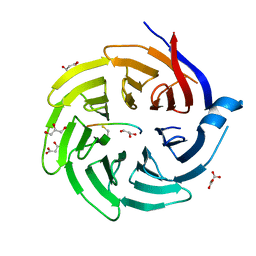 | |
3U5V
 
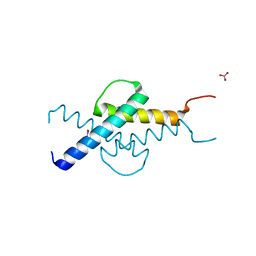 | | Crystal structure of Max-E47 | | Descriptor: | NITRATE ION, Protein max, Transcription factor E2-alpha chimera | | Authors: | Guarne, A, Ahmadpour, F, Gloyd, M. | | Deposit date: | 2011-10-11 | | Release date: | 2012-03-21 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of the minimalist max-e47 protein chimera.
Plos One, 7, 2012
|
|
5GNJ
 
 | | Structure of a transcription factor and DNA complex | | Descriptor: | DNA (5'-D(*AP*GP*GP*AP*AP*CP*AP*CP*GP*TP*GP*AP*CP*CP*C)-3'), DNA (5'-D(*TP*GP*GP*GP*TP*CP*AP*CP*GP*TP*GP*TP*TP*CP*C)-3'), Transcription factor MYC2 | | Authors: | Lian, T, Xu, Y, Su, X. | | Deposit date: | 2016-07-21 | | Release date: | 2017-05-10 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal Structure of Tetrameric Arabidopsis MYC2 Reveals the Mechanism of Enhanced Interaction with DNA.
Cell Rep, 19, 2017
|
|
5EYO
 
 | | The crystal structure of the Max bHLH domain in complex with 5-carboxyl cytosine DNA | | Descriptor: | DNA (5'-D(*AP*GP*TP*AP*GP*CP*AP*(1CC)P*GP*TP*GP*CP*TP*AP*CP*T)-3'), Protein max | | Authors: | Wang, D, Hashimoto, H, Zhang, X, Cheng, X. | | Deposit date: | 2015-11-25 | | Release date: | 2016-12-14 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | MAX is an epigenetic sensor of 5-carboxylcytosine and is altered in multiple myeloma.
Nucleic Acids Res., 45, 2017
|
|
2MH3
 
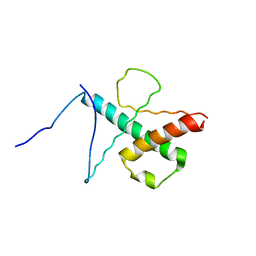 | | NMR structure of the basic helix-loop-helix region of the transcriptional repressor HES-1 | | Descriptor: | Transcription factor HES-1 | | Authors: | Wienk, H, Popovic, M, Coglievina, M, Boelens, R, Pongor, S, Pintar, A. | | Deposit date: | 2013-11-15 | | Release date: | 2014-02-05 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | The basic helix-loop-helix region of the transcriptional repressor hairy and enhancer of split 1 is preorganized to bind DNA.
Proteins, 82, 2014
|
|
2YPA
 
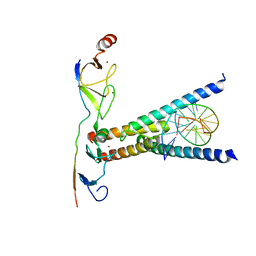 | | Structure of the SCL:E47:LMO2:LDB1 complex bound to DNA | | Descriptor: | EBOX FORWARD, EBOX REVERSE, LIM DOMAIN-BINDING PROTEIN 1, ... | | Authors: | El Omari, K, Hoosdally, S.J, Tuladhar, K, Karia, D, Ponsele, E, Platonova, O, Vyas, P, Patient, R, Porcher, C, Mancini, E.J. | | Deposit date: | 2012-10-30 | | Release date: | 2013-07-31 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural Basis for Lmo2-Driven Recruitment of the Scl:E47bHLH Heterodimer to Hematopoietic-Specific Transcriptional Targets.
Cell Rep., 4, 2013
|
|
2YPB
 
 | | Structure of the SCL:E47 complex bound to DNA | | Descriptor: | EBOX FORWARD, EBOX REVERSE, T-CELL ACUTE LYMPHOCYTIC LEUKEMIA PROTEIN 1, ... | | Authors: | El Omari, K, Hoosdally, S.J, Tuladhar, K, Karia, D, Ponsele, E, Platonova, O, Vyas, P, Patient, R, Porcher, C, Mancini, E.J. | | Deposit date: | 2012-10-30 | | Release date: | 2013-07-31 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.87 Å) | | Cite: | Structural Basis for Lmo2-Driven Recruitment of the Scl:E47bHLH Heterodimer to Hematopoietic-Specific Transcriptional Targets.
Cell Rep., 4, 2013
|
|
2A24
 
 | | HADDOCK Structure of HIF-2a/ARNT PAS-B Heterodimer | | Descriptor: | Aryl hydrocarbon receptor nuclear translocator, Endothelial PAS domain protein 1 | | Authors: | Card, P.B, Erbel, P.J, Gardner, K.H. | | Deposit date: | 2005-06-21 | | Release date: | 2006-01-17 | | Last modified: | 2022-03-09 | | Method: | SOLUTION NMR | | Cite: | Structural basis of ARNT PAS-B dimerization: use of a common beta-sheet interface for hetero- and homodimerization.
J.Mol.Biol., 353, 2005
|
|
1X0O
 
 | | human ARNT C-terminal PAS domain | | Descriptor: | Aryl hydrocarbon receptor nuclear translocator | | Authors: | Card, P.B, Erbel, P.J, Gardner, K.H. | | Deposit date: | 2005-03-25 | | Release date: | 2005-10-25 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | Structural Basis of ARNT PAS-B Dimerization: Use of a Common Beta-sheet Interface for Hetero- and Homodimerization.
J.Mol.Biol., 353, 2005
|
|
5Y7Y
 
 | | Crystal structure of AhRR/ARNT complex | | Descriptor: | Aryl hydrocarbon receptor nuclear translocator, Aryl hydrocarbon receptor repressor, GLYCEROL | | Authors: | Sakurai, S, Shimizu, T, Ohto, U. | | Deposit date: | 2017-08-18 | | Release date: | 2017-09-20 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The crystal structure of the AhRR-ARNT heterodimer reveals the structural basis of the repression of AhR-mediated transcription.
J. Biol. Chem., 292, 2017
|
|
4M4X
 
 | |
5NJ8
 
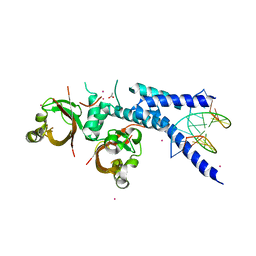 | |
7W80
 
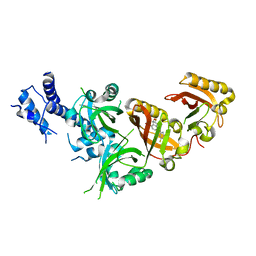 | | Crystal Structure of the Heterodimeric HIF-2 in Complex with Antagonist Belzutifan | | Descriptor: | 3-{[(1S,2S,3R)-2,3-difluoro-1-hydroxy-7-(methylsulfonyl)-2,3-dihydro-1H-inden-4-yl]oxy}-5-fluorobenzonitrile, Aryl hydrocarbon receptor nuclear translocator, Endothelial PAS domain-containing protein 1 | | Authors: | Ren, X, Diao, X, Zhuang, J, Wu, D. | | Deposit date: | 2021-12-07 | | Release date: | 2022-09-21 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.754 Å) | | Cite: | Structural basis for the allosteric inhibition of hypoxia-inducible factor (HIF)-2 by belzutifan.
Mol.Pharmacol., 2022
|
|
8T2I
 
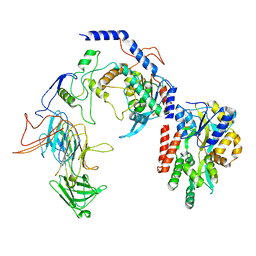 | | Negative stain EM assembly of MYC, JAZ, and NINJA complex | | Descriptor: | AFP homolog 2, Maltose/maltodextrin-binding periplasmic protein, Protein TIFY 10A, ... | | Authors: | Zhou, X.E, Zhang, Y, Zhou, Y, He, Q, Cao, X, Kariapper, L, Suino-Powell, K, Zhu, Y, Zhang, F, Karsten, M. | | Deposit date: | 2023-06-06 | | Release date: | 2023-06-28 | | Last modified: | 2023-11-22 | | Method: | ELECTRON MICROSCOPY (10.4 Å) | | Cite: | Assembly of JAZ-JAZ and JAZ-NINJA complexes in jasmonate signaling.
Plant Commun., 4, 2023
|
|
2K7S
 
 | |
4RQW
 
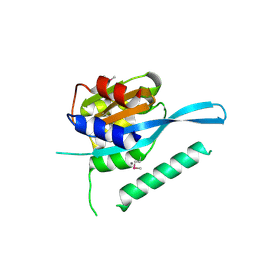 | | Crystal structure of Myc3 N-terminal JAZ-binding domain [44-238] from Arabidopsis | | Descriptor: | CALCIUM ION, Transcription factor MYC3 | | Authors: | Ke, J, Zhang, F, Zhou, X.E, Brunzelle, J, Zhou, M, Xu, H.E, Melcher, K, He, S.Y. | | Deposit date: | 2014-11-05 | | Release date: | 2015-08-12 | | Last modified: | 2015-09-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis of JAZ repression of MYC transcription factors in jasmonate signalling.
Nature, 525, 2015
|
|
4RRU
 
 | | Myc3 N-terminal JAZ-binding domain[5-242] from arabidopsis | | Descriptor: | CALCIUM ION, Transcription factor MYC3 | | Authors: | Ke, J, Zhang, F, Zhou, X.E, Brunzelle, J.S, Zhou, M, Xu, H.E, Melcher, K, He, S.Y. | | Deposit date: | 2014-11-06 | | Release date: | 2015-08-12 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis of JAZ repression of MYC transcription factors in jasmonate signalling.
Nature, 525, 2015
|
|
4RS9
 
 | | Structure of Myc3 N-terminal JAZ-binding domain [44-238] in complex with Jas motif of JAZ9 | | Descriptor: | Protein TIFY 7, Transcription factor MYC3 | | Authors: | Ke, J, Zhang, F, Zhou, X.E, Brunzelle, J.S, Zhou, M, Xu, H.E, Melcher, K, He, S.Y. | | Deposit date: | 2014-11-07 | | Release date: | 2015-08-12 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural basis of JAZ repression of MYC transcription factors in jasmonate signalling.
Nature, 525, 2015
|
|
3AY5
 
 | |
5T0Q
 
 | | Crystal structure of the Myc3 N-terminal domain [44-242] in complex with JAZ10 Jas domain [166-192] from arabidopsis | | Descriptor: | Protein TIFY 9, Transcription factor MYC3 | | Authors: | Ke, J, Zhang, F, Brunzelle, J.S, He, S.Y, Xu, H.E, Melcher, K. | | Deposit date: | 2016-08-16 | | Release date: | 2017-01-25 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structural insights into alternative splicing-mediated desensitization of jasmonate signaling.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5T0F
 
 | | Crystal structure of the Myc3 N-terminal domain [44-242] in complex with JAZ10 CMID domain [16-58] from arabidopsis | | Descriptor: | Protein TIFY 9, Transcription factor MYC3 | | Authors: | Ke, J, Zhang, F, Brunzelle, J.S, He, S.Y, Xu, H.E, Melcher, K. | | Deposit date: | 2016-08-16 | | Release date: | 2017-01-25 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural insights into alternative splicing-mediated desensitization of jasmonate signaling.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5T5X
 
 | |
4YWC
 
 | | Crystal structure of Myc3(5-242) fragment in complex with Jaz9(218-239) peptide | | Descriptor: | Protein TIFY 7, Transcription factor MYC3 | | Authors: | Ke, J, Zhang, F, Brunzelle, J.S, Xu, H.E, Melcher, K, He, S.Y. | | Deposit date: | 2015-03-20 | | Release date: | 2015-08-05 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis of JAZ repression of MYC transcription factors in jasmonate signalling.
Nature, 525, 2015
|
|
4YZ6
 
 | | Crystal Structure of Myc3[44-238] from Arabidopsis in complex with Jaz1 peptide [200-221] | | Descriptor: | Protein TIFY 10A, Transcription factor MYC3 | | Authors: | Ke, J, Zhang, F, Brunzelle, J, Xu, H.E, Melcher, K, HE, S.Y. | | Deposit date: | 2015-03-24 | | Release date: | 2015-08-05 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural basis of JAZ repression of MYC transcription factors in jasmonate signalling.
Nature, 525, 2015
|
|
8OSB
 
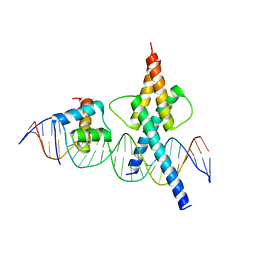 | | TWIST1-TCF4-ALX4 complex on specific DNA | | Descriptor: | DNA (25-MER), Homeobox protein aristaless-like 4, Transcription factor 4, ... | | Authors: | Morgunova, E, Kim, S, Popov, A, Wysocka, J, Taipale, J. | | Deposit date: | 2023-04-18 | | Release date: | 2024-01-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | DNA-guided transcription factor cooperativity shapes face and limb mesenchyme.
Cell, 187, 2024
|
|
