1O9B
 
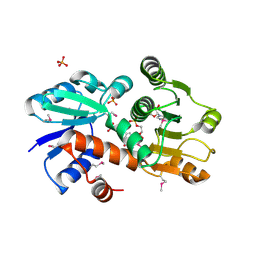 | | QUINATE/SHIKIMATE DEHYDROGENASE YDIB COMPLEXED WITH NADH | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, HYPOTHETICAL SHIKIMATE 5-DEHYDROGENASE-LIKE PROTEIN YDIB, PHOSPHATE ION | | Authors: | Michel, G, Cygler, M. | | Deposit date: | 2002-12-12 | | Release date: | 2003-02-01 | | Last modified: | 2013-10-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structures of Shikimate Dehydrogenase Aroe and its Paralog Ydib: A Common Structural Framework for Different Activities
J.Biol.Chem., 278, 2003
|
|
5SWV
 
 | |
6HQV
 
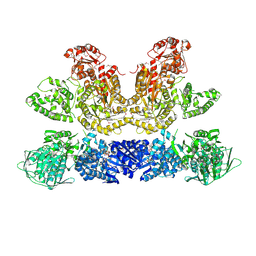 | | Pentafunctional AROM Complex from Chaetomium thermophilum | | Descriptor: | (3R,4S,5R)-3,4,5-TRIHYDROXYCYCLOHEX-1-ENE-1-CARBOXYLIC ACID, (4S,5R)-4,5-dihydroxy-3-oxocyclohex-1-ene-1-carboxylic acid, GLUTAMIC ACID, ... | | Authors: | Arora Verasto, H, Hartmann, M.D. | | Deposit date: | 2018-09-25 | | Release date: | 2020-05-06 | | Last modified: | 2020-08-26 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Architecture and functional dynamics of the pentafunctional AROM complex.
Nat.Chem.Biol., 16, 2020
|
|
6BMB
 
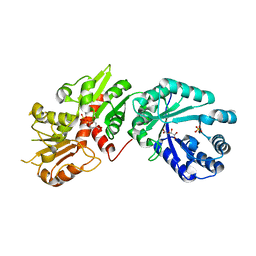 | | Crystal structure of Arabidopsis Dehydroquinate dehydratase-shikimate dehydrogenase (T381G mutant) in complex with tartrate and shikimate | | Descriptor: | (3R,4S,5R)-3,4,5-TRIHYDROXYCYCLOHEX-1-ENE-1-CARBOXYLIC ACID, Bifunctional 3-dehydroquinate dehydratase/shikimate dehydrogenase, chloroplastic, ... | | Authors: | Christendat, D, Peek, J. | | Deposit date: | 2017-11-14 | | Release date: | 2017-12-20 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.077 Å) | | Cite: | Structural and biochemical approaches uncover multiple evolutionary trajectories of plant quinate dehydrogenases.
Plant J., 2018
|
|
6BMQ
 
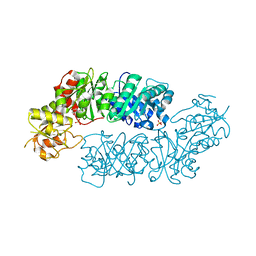 | | Crystal structure of Arabidopsis Dehydroquinate dehydratase-shikimate dehydrogenase (T381G mutant) in complex with tartrate and shikimate | | Descriptor: | (1S,3R,4S,5R)-1,3,4,5-tetrahydroxycyclohexanecarboxylic acid, Bifunctional 3-dehydroquinate dehydratase/shikimate dehydrogenase, chloroplastic, ... | | Authors: | Christendat, D, Peek, J. | | Deposit date: | 2017-11-15 | | Release date: | 2018-09-26 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.077 Å) | | Cite: | Structural and biochemical approaches uncover multiple evolutionary trajectories of plant quinate dehydrogenases.
Plant J., 2018
|
|
1WXD
 
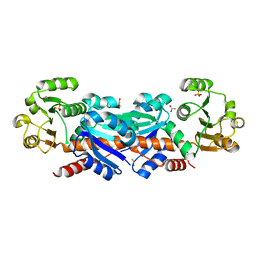 | |
2O7Q
 
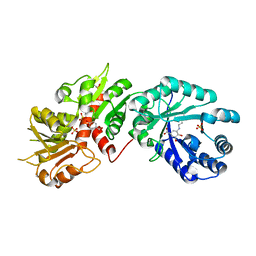 | |
2O7S
 
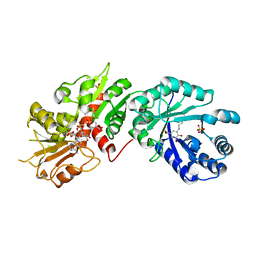 | |
3DOO
 
 | | Crystal structure of shikimate dehydrogenase from Staphylococcus epidermidis complexed with shikimate | | Descriptor: | (3R,4S,5R)-3,4,5-TRIHYDROXYCYCLOHEX-1-ENE-1-CARBOXYLIC ACID, Shikimate dehydrogenase | | Authors: | Han, C, Hu, T, Wu, D, Zhou, J, Shen, X, Qu, D, Jiang, H. | | Deposit date: | 2008-07-05 | | Release date: | 2009-05-05 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | X-ray crystallographic and enzymatic analyses of shikimate dehydrogenase from Staphylococcus epidermidis
Febs J., 276, 2009
|
|
3DON
 
 | | Crystal structure of shikimate dehydrogenase from Staphylococcus epidermidis | | Descriptor: | GLYCEROL, Shikimate dehydrogenase | | Authors: | Han, C, Hu, T, Wu, D, Zhou, J, Shen, X, Qu, D, Jiang, H. | | Deposit date: | 2008-07-05 | | Release date: | 2009-05-05 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | X-ray crystallographic and enzymatic analyses of shikimate dehydrogenase from Staphylococcus epidermidis
Febs J., 276, 2009
|
|
2CY0
 
 | |
2D5C
 
 | | Crystal Structure of Shikimate 5-Dehydrogenase (AroE) from Thermus Thermophilus HB8 in complex with shikimate | | Descriptor: | (3R,4S,5R)-3,4,5-TRIHYDROXYCYCLOHEX-1-ENE-1-CARBOXYLIC ACID, SULFATE ION, shikimate 5-dehydrogenase | | Authors: | Bagautdinov, B, Kunishima, N, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-10-31 | | Release date: | 2006-05-01 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal Structures of Shikimate Dehydrogenase AroE from Thermus thermophilus HB8 and its Cofactor and Substrate Complexes: Insights into the Enzymatic Mechanism
J.Mol.Biol., 373, 2007
|
|
3FBT
 
 | | Crystal structure of a chorismate mutase/shikimate 5-dehydrogenase fusion protein from Clostridium acetobutylicum | | Descriptor: | SULFATE ION, chorismate mutase and shikimate 5-dehydrogenase fusion protein | | Authors: | Bonanno, J.B, Gilmore, M, Bain, K.T, Hu, S, Romero, R, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-11-19 | | Release date: | 2008-12-09 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of a chorismate mutase/shikimate 5-dehydrogenase fusion protein from Clostridium acetobutylicum
To be Published
|
|
2EGG
 
 | |
7TBV
 
 | | Crystal structure of the shikimate kinase + 3-dehydroquinate dehydratase + 3-dehydroshikimate dehydrogenase domains of Aro1 from Candida albicans | | Descriptor: | CHLORIDE ION, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Stogios, P.J, Evdokimova, E, Michalska, K, Di Leo, R, Savchenko, A, Joachimiak, A, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2021-12-22 | | Release date: | 2022-03-16 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Molecular analysis and essentiality of Aro1 shikimate biosynthesis multi-enzyme in Candida albicans.
Life Sci Alliance, 5, 2022
|
|
7U5U
 
 | |
7U5T
 
 | |
7U5S
 
 | |
4OMU
 
 | |
2EV9
 
 | | Crystal Structure of Shikimate 5-Dehydrogenase (AroE) from Thermus Thermophilus HB8 in complex with NADP(H) and shikimate | | Descriptor: | (3R,4S,5R)-3,4,5-TRIHYDROXYCYCLOHEX-1-ENE-1-CARBOXYLIC ACID, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, SULFATE ION, ... | | Authors: | Bagautdinov, B, Kunishima, N, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-10-31 | | Release date: | 2006-05-01 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structures of Shikimate Dehydrogenase AroE from Thermus thermophilus HB8 and its Cofactor and Substrate Complexes: Insights into the Enzymatic Mechanism
J.Mol.Biol., 373, 2007
|
|
3SEF
 
 | | 2.4 Angstrom resolution crystal structure of shikimate 5-dehydrogenase (aroE) from Vibrio cholerae O1 biovar eltor str. N16961 in complex with shikimate and NADPH | | Descriptor: | (3R,4S,5R)-3,4,5-TRIHYDROXYCYCLOHEX-1-ENE-1-CARBOXYLIC ACID, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Shikimate 5-dehydrogenase | | Authors: | Halavaty, A.S, Light, S.H, Minasov, G, Shuvalova, L, Papazisi, L, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-06-10 | | Release date: | 2011-07-27 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | 2.4 Angstrom resolution crystal structure of shikimate 5-dehydrogenase (aroE) from Vibrio cholerae O1 biovar eltor str. N16961 in complex with shikimate and NADPH
To be Published
|
|
2GPT
 
 | | Crystal structure of Arabidopsis Dehydroquinate dehydratase-shikimate dehydrogenase in complex with tartrate and shikimate | | Descriptor: | (3R,4S,5R)-3,4,5-TRIHYDROXYCYCLOHEX-1-ENE-1-CARBOXYLIC ACID, 3-dehydroquinate dehydratase/ shikimate 5-dehydrogenase, L(+)-TARTARIC ACID, ... | | Authors: | Singh, S.A, Christendat, D. | | Deposit date: | 2006-04-18 | | Release date: | 2006-06-06 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure of Arabidopsis dehydroquinate dehydratase-shikimate dehydrogenase and implications for metabolic channeling in the shikimate pathway
Biochemistry, 45, 2006
|
|
2HK9
 
 | | Crystal structure of shikimate dehydrogenase from aquifex aeolicus in complex with shikimate and NADP+ at 2.2 angstrom resolution | | Descriptor: | (3R,4S,5R)-3,4,5-TRIHYDROXYCYCLOHEX-1-ENE-1-CARBOXYLIC ACID, 2'-MONOPHOSPHOADENOSINE-5'-DIPHOSPHATE, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Gan, J.H, Prabakaran, P, Gu, Y.J, Andrykovitch, M, Li, Y, Liu, H.H, Yan, H, Ji, X. | | Deposit date: | 2006-07-03 | | Release date: | 2007-06-19 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural and biochemical analyses of shikimate dehydrogenase AroE from Aquifex aeolicus: implications for the catalytic mechanism.
Biochemistry, 46, 2007
|
|
2HK8
 
 | | Crystal structure of shikimate dehydrogenase from aquifex aeolicus at 2.35 angstrom resolution | | Descriptor: | Shikimate dehydrogenase | | Authors: | Gan, J.H, Prabakaran, P, Gu, Y.J, Andrykovitch, M, Li, Y, Liu, H.H, Yan, H, Ji, X. | | Deposit date: | 2006-07-03 | | Release date: | 2007-06-19 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural and biochemical analyses of shikimate dehydrogenase AroE from Aquifex aeolicus: implications for the catalytic mechanism.
Biochemistry, 46, 2007
|
|
2HK7
 
 | | Crystal structure of shikimate dehydrogenase from aquifex aeolicus in complex with mercury at 2.5 angstrom resolution | | Descriptor: | MERCURY (II) ION, Shikimate dehydrogenase | | Authors: | Gan, J.H, Prabakaran, P, Gu, Y.J, Andrykovitch, M, Li, Y, Liu, H.H, Yan, H, Ji, X. | | Deposit date: | 2006-07-03 | | Release date: | 2007-06-19 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural and biochemical analyses of shikimate dehydrogenase AroE from Aquifex aeolicus: implications for the catalytic mechanism.
Biochemistry, 46, 2007
|
|
