4GF9
 
 | | Structural insights into the dual strategy of recognition of peptidoglycan recognition protein, PGRP-S: ternary complex of PGRP-S with LPS and fatty acid | | Descriptor: | (R)-((2R,3S,4R,5R,6R)-3-HYDROXY-2-(HYDROXYMETHYL)-5-((R)-3-HYDROXYTETRADECANAMIDO)-6-(PHOSPHONOOXY)TETRAHYDRO-2H-PYRAN-4-YL) 3-HYDROXYTETRADECANOATE, GLYCEROL, Peptidoglycan recognition protein 1, ... | | Authors: | Sharma, P, Dube, D, Sinha, M, Yadav, S, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2012-08-03 | | Release date: | 2012-09-26 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural insights into the dual strategy of recognition by peptidoglycan recognition protein, PGRP-S: structure of the ternary complex of PGRP-S with lipopolysaccharide and stearic acid.
Plos One, 8, 2013
|
|
5E0B
 
 | | Crystal structure of the complex of Peptidoglycan recognition protein PGRP-S with N-Acetyl Muramic acid at 2.6 A resolution | | Descriptor: | GLYCEROL, L(+)-TARTARIC ACID, N-acetyl-beta-muramic acid, ... | | Authors: | Sharma, P, Yamini, S, Dube, D, Sinha, M, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2015-09-28 | | Release date: | 2015-10-14 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of the complex of Peptidoglycan recognition protein PGRP-S with N-Acetyl Muramic acid at 2.6 A resolution
To Be Published
|
|
5E0A
 
 | | Crystal Structure of the complex of Camel Peptidoglycan Recognition Protein (CPGRP-S) and N-Acetylglucosamine at 2.6 A | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, L(+)-TARTARIC ACID, Peptidoglycan recognition protein 1 | | Authors: | Dube, D, Sharma, P, Sinha, M, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2015-09-28 | | Release date: | 2015-10-14 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal Structure of the complex of Camel Peptidoglycan Recognition Protein (CPGRP-S) and N-Acetylglucosamine at 2.6 A
To Be Published
|
|
7XFX
 
 | | Crystal structure of the ternary complex of Peptidoglycan recognition protein, PGRP-S with hexanoic and tartaric acids at 2.28 A resolution. | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, CHLORIDE ION, ... | | Authors: | Maurya, A, Singh, P.K, Viswanathan, V, Sharma, P, Sharma, S, Singh, T.P. | | Deposit date: | 2022-04-02 | | Release date: | 2022-05-11 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Crystal structure of the ternary complex of Peptidoglycan recognition protein, PGRP-S with hexanoic and tartaric acids at 2.28 A resolution.
To Be Published
|
|
7XFY
 
 | | Crystal structure of the ternary complex of Peptidoglycan recognition protein, PGRP-S with hexanoic and tartaric acids at 2.67 A resolution. | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, CHLORIDE ION, ... | | Authors: | Maurya, A, Singh, P.K, Viswanathan, V, Sharma, P, Sharma, S, Singh, T.P. | | Deposit date: | 2022-04-02 | | Release date: | 2022-05-11 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.67 Å) | | Cite: | Crystal structure of the ternary complex of Peptidoglycan recognition protein, PGRP-S with hexanoic and tartaric acids at 2.67 A resolution.
To Be Published
|
|
7XFW
 
 | | Crystal structure of the ternary complex of Peptidoglycan recognition protein, PGRP-S with hexanoic and tartaric acids at 2.07 A resolution. | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, CHLORIDE ION, ... | | Authors: | Maurya, A, Singh, P.K, Viswanathan, V, Sharma, P, Sharma, S, Singh, T.P. | | Deposit date: | 2022-04-02 | | Release date: | 2022-05-11 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Structure of the complex of camel peptidoglycan recognition protein-S with hexanoic acid reveals novel features of the versatile ligand-binding site at the dimeric interface.
Biochim Biophys Acta Proteins Proteom, 1871, 2022
|
|
7XU8
 
 | | Structure of the complex of camel peptidoglycan recognition protein-short (PGRP-S) with heptanoic acid at 2.15 A resolution. | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1,2-ETHANEDIOL, CARBONATE ION, ... | | Authors: | Maurya, A, Ahmad, N, Viswanathan, V, Singh, P.K, Yamini, S, Sharma, P, Sinha, M, Bhushan, A, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2022-05-18 | | Release date: | 2022-06-15 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Ligand recognition by peptidoglycan recognition protein-S (PGRP-S): structure of the complex of camel PGRP-S with heptanoic acid at 2.15 angstrom resolution.
Int J Biochem Mol Biol, 13, 2022
|
|
7NSZ
 
 | | Drosophila PGRP-LB Y78F mutant | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Isoform A of Peptidoglycan-recognition protein LB, SODIUM ION, ... | | Authors: | Orlans, J, Aller, P, Da Silva, P. | | Deposit date: | 2021-03-08 | | Release date: | 2021-05-19 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | PGRP-LB: An Inside View into the Mechanism of the Amidase Reaction.
Int J Mol Sci, 22, 2021
|
|
7NSY
 
 | | Drosophila PGRP-LB C160S mutant | | Descriptor: | Isoform A of Peptidoglycan-recognition protein LB | | Authors: | Orlans, J, Aller, P, Da Silva, P. | | Deposit date: | 2021-03-08 | | Release date: | 2021-05-19 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | PGRP-LB: An Inside View into the Mechanism of the Amidase Reaction.
Int J Mol Sci, 22, 2021
|
|
7NT0
 
 | | Drosophila PGRP-LB Y78F mutant in complex with tracheal cytotoxin (TCT) | | Descriptor: | GLCNAC(BETA1-4)-MURNAC(1,6-ANHYDRO)-L-ALA-GAMMA-D-GLU-MESO-A2PM-D-ALA, Isoform A of Peptidoglycan-recognition protein LB, ZINC ION | | Authors: | Orlans, J, Aller, P, Da Silva, P. | | Deposit date: | 2021-03-08 | | Release date: | 2021-05-19 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | PGRP-LB: An Inside View into the Mechanism of the Amidase Reaction.
Int J Mol Sci, 22, 2021
|
|
7NSX
 
 | | Drosophila PGRP-LB wild-type | | Descriptor: | Isoform A of Peptidoglycan-recognition protein LB, ZINC ION | | Authors: | Orlans, J, Aller, P, Da Silva, P. | | Deposit date: | 2021-03-08 | | Release date: | 2021-05-19 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | PGRP-LB: An Inside View into the Mechanism of the Amidase Reaction.
Int J Mol Sci, 22, 2021
|
|
4IVV
 
 | |
1YCK
 
 | |
1YB0
 
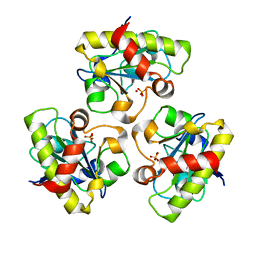 | | Structure of PlyL | | Descriptor: | PHOSPHATE ION, ZINC ION, prophage LambdaBa02, ... | | Authors: | Low, L.Y, Yang, C, Perego, M, Osterman, A, Liddington, R.C. | | Deposit date: | 2004-12-18 | | Release date: | 2005-08-23 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Structure and lytic activity of a Bacillus anthracis prophage endolysin
J.Biol.Chem., 280, 2005
|
|
4KNL
 
 | |
4KNK
 
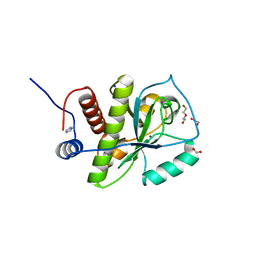 | | Crystal structure of Staphylococcus aureus hydrolase AmiA | | Descriptor: | 1,2-ETHANEDIOL, Bifunctional autolysin, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Buettner, F.M, Zoll, S, Stehle, T. | | Deposit date: | 2013-05-10 | | Release date: | 2014-03-12 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.124 Å) | | Cite: | Structure-function analysis of Staphylococcus aureus amidase reveals the determinants of peptidoglycan recognition and cleavage.
J.Biol.Chem., 289, 2014
|
|
1Z6I
 
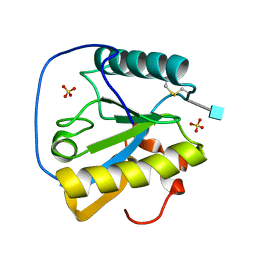 | | Crystal structure of the ectodomain of Drosophila transmembrane receptor PGRP-LCa | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Peptidoglycan-recognition protein-LC, SULFATE ION | | Authors: | Chang, C.-I, Ihara, K, Chelliah, Y, Mengin-Lecreulx, D, Wakatsuki, S, Deisenhofer, J. | | Deposit date: | 2005-03-22 | | Release date: | 2005-07-19 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of the ectodomain of Drosophila peptidoglycan-recognition protein LCa suggests a molecular mechanism for pattern recognition
Proc.Natl.Acad.Sci.Usa, 102, 2005
|
|
1OHT
 
 | | Peptidoglycan recognition protein LB | | Descriptor: | 1,2-ETHANEDIOL, CG14704 PROTEIN, L(+)-TARTARIC ACID, ... | | Authors: | Kim, M.-S, Byun, M, Oh, B.-H. | | Deposit date: | 2003-05-31 | | Release date: | 2003-07-18 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of Peptidoglycan Recognition Protein Lb from Drosophila Melanogaster
Nat.Immunol., 4, 2003
|
|
1S2J
 
 | | Crystal structure of the Drosophila pattern-recognition receptor PGRP-SA | | Descriptor: | PHOSPHATE ION, Peptidoglycan recognition protein SA CG11709-PA | | Authors: | Chang, C.-I, Pili-Floury, S, Chelliah, Y, Lemaitre, B, Mengin-Lecreulx, D, Deisenhofer, J. | | Deposit date: | 2004-01-08 | | Release date: | 2004-09-14 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | A Drosophila pattern recognition receptor contains a peptidoglycan docking groove and unusual l,d-carboxypeptidase activity.
PLOS BIOL., 2, 2004
|
|
1SK3
 
 | | Crystal structure of the C-terminal peptidoglycan-binding domain of human peptidoglycan recognition protein Ialpha | | Descriptor: | NICKEL (II) ION, Peptidoglycan recognition protein I-alpha, SULFATE ION | | Authors: | Guan, R, Malchiodi, E.L, Qian, W, Schuck, P, Mariuzza, R.A. | | Deposit date: | 2004-03-04 | | Release date: | 2004-07-13 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of the C-terminal peptidoglycan-binding domain of human peptidoglycan recognition protein Ialpha
J.Biol.Chem., 279, 2004
|
|
1SK4
 
 | | crystal structure of the C-terminal peptidoglycan-binding domain of human peptidoglycan recognition protein Ialpha | | Descriptor: | Peptidoglycan recognition protein I-alpha, SODIUM ION | | Authors: | Guan, R, Malchiodi, E.L, Qian, W, Schuck, P, Mariuzza, R.A. | | Deposit date: | 2004-03-04 | | Release date: | 2004-07-13 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal structure of the C-terminal peptidoglycan-binding domain of human peptidoglycan recognition protein Ialpha
J.Biol.Chem., 279, 2004
|
|
1SXR
 
 | | Drosophila Peptidoglycan Recognition Protein (PGRP)-SA | | Descriptor: | 1,2-ETHANEDIOL, Peptidoglycan recognition protein SA CG11709-PA, SULFATE ION | | Authors: | Reiser, J.B, Teyton, L, Wilson, I.A. | | Deposit date: | 2004-03-31 | | Release date: | 2004-06-29 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Crystal structure of the Drosophila peptidoglycan recognition protein (PGRP)-SA at 1.56 A resolution
J.Mol.Biol., 340, 2004
|
|
3T2V
 
 | | Crystal structure of the complex of peptidoglycan recognition protein-short (CPGRP-S) with mycolic acid at 2.5 A resolution | | Descriptor: | (2S,3R)-2-hexyl-3-hydroxynonanoic acid, GLYCEROL, L(+)-TARTARIC ACID, ... | | Authors: | Sharma, P, Dube, D, Sinha, M, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2011-07-23 | | Release date: | 2011-08-10 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Structural basis of the binding of fatty acids to peptidoglycan recognition protein, PGRP-S through second binding site
Arch.Biochem.Biophys., 529, 2013
|
|
3T39
 
 | | Crystal structure of the complex of camel peptidoglycan recognition protein(CPGRP-S) with a mycobacterium metabolite shikimate at 2.7 A resolution | | Descriptor: | (3R,4S,5R)-3,4,5-TRIHYDROXYCYCLOHEX-1-ENE-1-CARBOXYLIC ACID, GLYCEROL, Peptidoglycan recognition protein 1, ... | | Authors: | Sharma, P, Dube, D, Sinha, M, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2011-07-25 | | Release date: | 2011-08-24 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of the complex of peptidoglycan recognition protein-short (CPGRP-S) with a mycobacterium metabolite shikimate at 2.7 A resolution
To be Published
|
|
3TRU
 
 | | Crystal structure of the complex of peptidoglycan recognition protein with cellular metabolite chorismate at 3.2 A resolution | | Descriptor: | (3R,4R)-3-[(1-carboxyethenyl)oxy]-4-hydroxycyclohexa-1,5-diene-1-carboxylic acid, GLYCEROL, L(+)-TARTARIC ACID, ... | | Authors: | Dube, D, Sharma, P, Sinha, M, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2011-09-10 | | Release date: | 2011-09-28 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Crystal structure of the complex of peptidoglycan recognition protein with cellular metabolite chorismate at 3.2 A resolution
To be Published
|
|
