6G2D
 
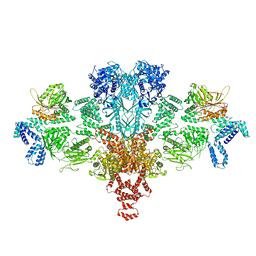 | | Citrate-induced acetyl-CoA carboxylase (ACC-Cit) filament at 5.4 A resolution | | Descriptor: | Acetyl-CoA carboxylase 1 | | Authors: | Hunkeler, M, Hagmann, A, Stuttfeld, E, Chami, M, Stahlberg, H, Maier, T. | | Deposit date: | 2018-03-23 | | Release date: | 2018-06-13 | | Last modified: | 2019-12-11 | | Method: | ELECTRON MICROSCOPY (5.4 Å) | | Cite: | Structural basis for regulation of human acetyl-CoA carboxylase.
Nature, 558, 2018
|
|
7KCT
 
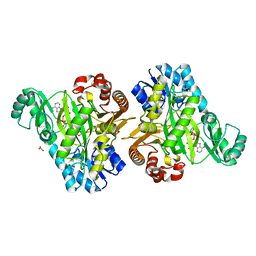 | | Crystal Structure of the Hydrogenobacter thermophilus 2-Oxoglutarate Carboxylase (OGC) Biotin Carboxylase (BC) Domain Dimer in Complex with Adenosine 5'-Diphosphate Magnesium Salt (MgADP), Adenosine 5'-Diphosphate (ADP, and Bicarbonate Anion (Hydrogen Carbonate/HCO3-) | | Descriptor: | 2-oxoglutarate carboxylase small subunit, ADENOSINE-5'-DIPHOSPHATE, BICARBONATE ION, ... | | Authors: | Buhrman, G.K, Rose, R.B, Enriquez, P, Truong, V. | | Deposit date: | 2020-10-07 | | Release date: | 2021-01-27 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Structure, Function, and Thermal Adaptation of the Biotin Carboxylase Domain Dimer from Hydrogenobacter thermophilus 2-Oxoglutarate Carboxylase.
Biochemistry, 60, 2021
|
|
7KC7
 
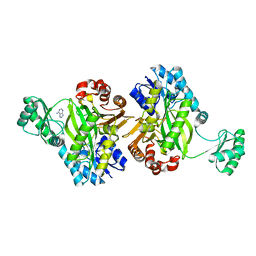 | | Biotin Carboxylase domain of Thermophilic 2-Oxoglutarate Carboxylase bound to ADP without Magnesium with disordered phosphate tail | | Descriptor: | 2-oxoglutarate carboxylase small subunit, ADENOSINE-5'-DIPHOSPHATE, PHOSPHATE ION | | Authors: | Buhrman, G.K, Rose, R.B, Enriquez, P, Truong, V. | | Deposit date: | 2020-10-05 | | Release date: | 2021-01-27 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure, Function, and Thermal Adaptation of the Biotin Carboxylase Domain Dimer from Hydrogenobacter thermophilus 2-Oxoglutarate Carboxylase.
Biochemistry, 60, 2021
|
|
7KBL
 
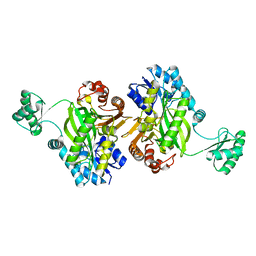 | | Biotin Carboxylase domain of Thermophilic 2-Oxoglutarate Carboxylase bound to Bicarbonate | | Descriptor: | 2-oxoglutarate carboxylase small subunit, BICARBONATE ION | | Authors: | Buhrman, G.K, Rose, R.B, Enriquez, P, Truong, V. | | Deposit date: | 2020-10-02 | | Release date: | 2021-01-27 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure, Function, and Thermal Adaptation of the Biotin Carboxylase Domain Dimer from Hydrogenobacter thermophilus 2-Oxoglutarate Carboxylase.
Biochemistry, 60, 2021
|
|
3GID
 
 | |
3G8C
 
 | | Crystal Structure of Biotin Carboxylase in Complex with Biotin, Bicarbonate, ADP and Mg Ion | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BICARBONATE ION, BIOTIN, ... | | Authors: | Chou, C.Y, Yu, L.P, Tong, L. | | Deposit date: | 2009-02-12 | | Release date: | 2009-03-03 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of biotin carboxylase in complex with substrates and implications for its catalytic mechanism.
J.Biol.Chem., 284, 2009
|
|
3G8D
 
 | | Crystal structure of the biotin carboxylase subunit, E296A mutant, of acetyl-COA carboxylase from Escherichia coli | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Biotin carboxylase, MAGNESIUM ION, ... | | Authors: | Chou, C.Y, Yu, L.P, Tong, L. | | Deposit date: | 2009-02-12 | | Release date: | 2009-03-03 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of biotin carboxylase in complex with substrates and implications for its catalytic mechanism.
J.Biol.Chem., 284, 2009
|
|
2GPW
 
 | | Crystal Structure of the Biotin Carboxylase Subunit, F363A Mutant, of Acetyl-CoA Carboxylase from Escherichia coli. | | Descriptor: | Biotin carboxylase | | Authors: | Shen, Y, Chou, C.Y, Chang, G.G, Tong, L. | | Deposit date: | 2006-04-18 | | Release date: | 2006-07-04 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Is dimerization required for the catalytic activity of bacterial biotin carboxylase?
Mol.Cell, 22, 2006
|
|
2GPS
 
 | | Crystal Structure of the Biotin Carboxylase Subunit, E23R mutant, of Acetyl-CoA Carboxylase from Escherichia coli. | | Descriptor: | Biotin carboxylase | | Authors: | Shen, Y, Chou, C.Y, Chang, G.G, Tong, L. | | Deposit date: | 2006-04-18 | | Release date: | 2006-07-04 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Is dimerization required for the catalytic activity of bacterial biotin carboxylase?
Mol.Cell, 22, 2006
|
|
2HJW
 
 | | Crystal Structure of the BC domain of ACC2 | | Descriptor: | Acetyl-CoA carboxylase 2 | | Authors: | Cho, Y.S, Lee, J.I, Shin, D, Kim, H.T, Lee, T.G, Heo, Y.S. | | Deposit date: | 2006-07-02 | | Release date: | 2007-07-03 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of the biotin carboxylase domain of human acetyl-CoA carboxylase 2.
Proteins, 70, 2008
|
|
2J9G
 
 | |
5H80
 
 | | Biotin Carboxylase domain of single-chain bacterial carboxylase | | Descriptor: | 1,2-ETHANEDIOL, Carboxylase | | Authors: | Hagmann, A, Hunkeler, M, Stuttfeld, E, Maier, T. | | Deposit date: | 2015-12-23 | | Release date: | 2016-07-20 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Hybrid Structure of a Dynamic Single-Chain Carboxylase from Deinococcus radiodurans.
Structure, 24, 2016
|
|
5I6I
 
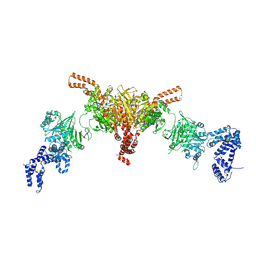 | | Crystal structure of a dBCCP-variant of Chaetomium thermophilum acetyl-CoA carboxylase | | Descriptor: | Acetyl-CoA carboxylase-like protein,Acetyl-CoA carboxylase-like protein,Acetyl-CoA carboxylase-like protein | | Authors: | Hunkeler, M, Stuttfeld, E, Hagmann, A, Imseng, S, Maier, T. | | Deposit date: | 2016-02-16 | | Release date: | 2016-04-20 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (8.4 Å) | | Cite: | The dynamic organization of fungal acetyl-CoA carboxylase.
Nat Commun, 7, 2016
|
|
5I8I
 
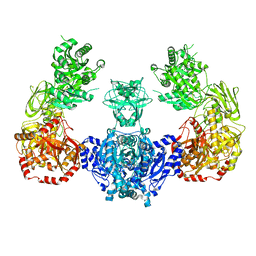 | |
3TW6
 
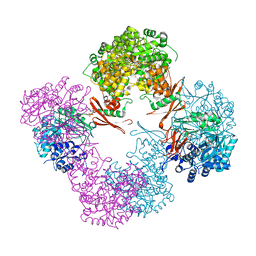 | | Structure of Rhizobium etli pyruvate carboxylase T882A with the allosteric activator, acetyl coenzyme-A | | Descriptor: | 5-(HEXAHYDRO-2-OXO-1H-THIENO[3,4-D]IMIDAZOL-6-YL)PENTANAL, ADENOSINE-5'-DIPHOSPHATE, CHLORIDE ION, ... | | Authors: | St Maurice, M, Kumar, S, Lietzan, A.D. | | Deposit date: | 2011-09-21 | | Release date: | 2011-10-19 | | Last modified: | 2011-12-28 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Interaction between the biotin carboxyl carrier domain and the biotin carboxylase domain in pyruvate carboxylase from Rhizobium etli.
Biochemistry, 50, 2011
|
|
3TW7
 
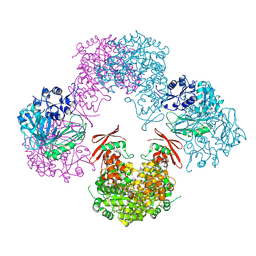 | | Structure of Rhizobium etli pyruvate carboxylase T882A crystallized without acetyl coenzyme-A | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, Pyruvate carboxylase protein, ... | | Authors: | St Maurice, M, Kumar, S, Lietzan, A.D. | | Deposit date: | 2011-09-21 | | Release date: | 2011-10-12 | | Last modified: | 2011-12-28 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Interaction between the biotin carboxyl carrier domain and the biotin carboxylase domain in pyruvate carboxylase from Rhizobium etli.
Biochemistry, 50, 2011
|
|
3U9T
 
 | |
4HR7
 
 | | Crystal Structure of Biotin Carboxyl Carrier Protein-Biotin Carboxylase Complex from E.coli | | Descriptor: | 1,2-ETHANEDIOL, Biotin carboxyl carrier protein of acetyl-CoA carboxylase, Biotin carboxylase, ... | | Authors: | Broussard, T.C, Kobe, M.J, Pakhomova, S, Neau, D.B, Price, A.E, Champion, T.S, Waldrop, G.L. | | Deposit date: | 2012-10-26 | | Release date: | 2013-03-13 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.495 Å) | | Cite: | The three-dimensional structure of the biotin carboxylase-biotin carboxyl carrier protein complex of E. coli acetyl-CoA carboxylase.
Structure, 21, 2013
|
|
3U9S
 
 | | Crystal structure of P. aeruginosa 3-methylcrotonyl-CoA carboxylase (MCC) 750 kD holoenzyme, CoA complex | | Descriptor: | 5-(HEXAHYDRO-2-OXO-1H-THIENO[3,4-D]IMIDAZOL-6-YL)PENTANAL, COENZYME A, Methylcrotonyl-CoA carboxylase, ... | | Authors: | Huang, C.S, Tong, L. | | Deposit date: | 2011-10-19 | | Release date: | 2011-12-14 | | Last modified: | 2013-01-23 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | An unanticipated architecture of the 750-kDa {alpha}6{beta}6 holoenzyme of 3-methylcrotonyl-CoA carboxylase
Nature, 481, 2012
|
|
4HNU
 
 | | crystal structure of K442E mutant of S. aureus Pyruvate carboxylase | | Descriptor: | 5-(HEXAHYDRO-2-OXO-1H-THIENO[3,4-D]IMIDAZOL-6-YL)PENTANAL, ADENOSINE-5'-DIPHOSPHATE, MANGANESE (II) ION, ... | | Authors: | Yu, L.P.C, Tong, L. | | Deposit date: | 2012-10-21 | | Release date: | 2013-01-16 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Characterizing the Importance of the Biotin Carboxylase Domain Dimer for Staphylococcus aureus Pyruvate Carboxylase Catalysis.
Biochemistry, 52, 2013
|
|
4HQ6
 
 | | BC domain in the presence of citrate | | Descriptor: | Acetyl-CoA carboxylase 2 | | Authors: | Heo, Y.S. | | Deposit date: | 2012-10-25 | | Release date: | 2013-10-09 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural Insights into the Regulation of ACC2 by Citrate
Bull.Korean Chem.Soc., 34, 2013
|
|
4HNV
 
 | | Crystal structure of R54E mutant of S. aureus Pyruvate carboxylase | | Descriptor: | 5-(HEXAHYDRO-2-OXO-1H-THIENO[3,4-D]IMIDAZOL-6-YL)PENTANAL, ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Yu, L.P.C, Tong, L. | | Deposit date: | 2012-10-21 | | Release date: | 2013-01-16 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Characterizing the Importance of the Biotin Carboxylase Domain Dimer for Staphylococcus aureus Pyruvate Carboxylase Catalysis.
Biochemistry, 52, 2013
|
|
4HNT
 
 | | crystal structure of F403A mutant of S. aureus Pyruvate carboxylase | | Descriptor: | 5-(HEXAHYDRO-2-OXO-1H-THIENO[3,4-D]IMIDAZOL-6-YL)PENTANAL, ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Yu, L.P.C, Tong, L. | | Deposit date: | 2012-10-21 | | Release date: | 2013-01-16 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Characterizing the Importance of the Biotin Carboxylase Domain Dimer for Staphylococcus aureus Pyruvate Carboxylase Catalysis.
Biochemistry, 52, 2013
|
|
3VA7
 
 | | Crystal structure of the Kluyveromyces lactis Urea Carboxylase | | Descriptor: | 5-(HEXAHYDRO-2-OXO-1H-THIENO[3,4-D]IMIDAZOL-6-YL)PENTANAL, GLYCEROL, KLLA0E08119p, ... | | Authors: | Fan, C, Xiang, S. | | Deposit date: | 2011-12-29 | | Release date: | 2012-02-01 | | Last modified: | 2013-07-17 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of urea carboxylase provides insights into the carboxyltransfer reaction
J.Biol.Chem., 287, 2012
|
|
5KKN
 
 | | Crystal structure of human ACC2 BC domain in complex with ND-646, the primary amide of ND-630 | | Descriptor: | 2-[1-[(2~{R})-2-(2-methoxyphenyl)-2-(oxan-4-yloxy)ethyl]-5-methyl-6-(1,3-oxazol-2-yl)-2,4-bis(oxidanylidene)thieno[2,3-d]pyrimidin-3-yl]-2-methyl-propanamide, Acetyl-CoA carboxylase 2 | | Authors: | Wang, R, Paul, D, Tong, L. | | Deposit date: | 2016-06-22 | | Release date: | 2016-07-13 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Acetyl-CoA carboxylase inhibition by ND-630 reduces hepatic steatosis, improves insulin sensitivity, and modulates dyslipidemia in rats.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
