1J58
 
 | | Crystal Structure of Oxalate Decarboxylase | | Descriptor: | FORMIC ACID, MAGNESIUM ION, MANGANESE (II) ION, ... | | Authors: | Anand, R, Dorrestein, P.C, Kinsland, C, Begley, T.P, Ealick, S.E. | | Deposit date: | 2002-02-25 | | Release date: | 2002-07-03 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structure of oxalate decarboxylase from Bacillus subtilis at 1.75 A resolution.
Biochemistry, 41, 2002
|
|
1L3J
 
 | | Crystal Structure of Oxalate Decarboxylase Formate Complex | | Descriptor: | FORMIC ACID, MAGNESIUM ION, MANGANESE (II) ION, ... | | Authors: | Anand, R, Dorrestein, P.C, Kinsland, C, Begley, T.P, Ealick, S.E. | | Deposit date: | 2002-02-27 | | Release date: | 2002-07-03 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of oxalate decarboxylase from Bacillus subtilis at 1.75 A resolution.
Biochemistry, 41, 2002
|
|
1UD1
 
 | | Crystal structure of proglycinin mutant C88S | | Descriptor: | Glycinin G1 | | Authors: | Utsumi, S, Adachi, M. | | Deposit date: | 2003-04-24 | | Release date: | 2004-02-03 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Crystal Structures and Structural Stabilities of the Disulfide Bond-Deficient Soybean Proglycinin Mutants C12G and C88S.
J.Agric.Food Chem., 51, 2003
|
|
6UFI
 
 | | W96Y Oxalate Decarboxylase (Bacillus subtilis) | | Descriptor: | CHLORIDE ION, Cupin domain-containing protein, GLYCEROL, ... | | Authors: | Pastore, A.J, Burg, M.J, Twahir, U.T, Bruner, S.D, Angerhofer, A. | | Deposit date: | 2019-09-24 | | Release date: | 2020-09-30 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Oxalate decarboxylase uses electron hole hopping for catalysis.
J.Biol.Chem., 297, 2021
|
|
1UCX
 
 | | Crystal structure of proglycinin C12G mutant | | Descriptor: | Glycinin G1 | | Authors: | Utsumi, S, Adachi, M. | | Deposit date: | 2003-04-24 | | Release date: | 2004-02-03 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Crystal Structures and Structural Stabilities of the Disulfide Bond-Deficient Soybean Proglycinin Mutants C12G and C88S.
J.Agric.Food Chem., 51, 2003
|
|
6TZP
 
 | | W96F Oxalate Decarboxylase (B. subtilis) | | Descriptor: | MANGANESE (II) ION, Oxalate decarboxylase | | Authors: | Pastore, A.J, Burg, M.J, Twahir, U.T, Bruner, S.D, Angerhofer, A. | | Deposit date: | 2019-08-12 | | Release date: | 2021-02-17 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Oxalate decarboxylase uses electron hole hopping for catalysis.
J.Biol.Chem., 297, 2021
|
|
1UIK
 
 | | Crystal structure of soybean beta-conglycinin alpha prime homotrimer | | Descriptor: | MAGNESIUM ION, alpha prime subunit of beta-conglycinin | | Authors: | Maruyama, Y, Maruyama, N, Mikami, B, Utsumi, S. | | Deposit date: | 2003-07-16 | | Release date: | 2004-07-16 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of the core region of the soybean beta-conglycinin alpha' subunit.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
1UIJ
 
 | | Crystal Structure Of Soybean beta-Conglycinin Beta Homotrimer (I122M/K124W) | | Descriptor: | beta subunit of beta conglycinin | | Authors: | Maruyama, N, Maruyama, Y, Tsuruki, T, Okuda, E, Yoshikawa, M, Utsumi, S. | | Deposit date: | 2003-07-16 | | Release date: | 2004-07-16 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Creation of soybean beta-conglycinin beta with strong phagocytosis-stimulating activity
BIOCHIM.BIOPHYS.ACTA, 1648, 2003
|
|
6V7J
 
 | | The C2221 crystal form of canavalin at 173 K | | Descriptor: | BENZOIC ACID, CALCIUM ION, Canavalin, ... | | Authors: | McPherson, A. | | Deposit date: | 2019-12-08 | | Release date: | 2020-02-19 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Binding of benzoic acid and anions within the cupin domains of the vicilin protein canavalin from jack bean (Canavalia ensiformis): Crystal structures.
Biochem.Biophys.Res.Commun., 524, 2020
|
|
6V7G
 
 | |
1UW8
 
 | | CRYSTAL STRUCTURE OF OXALATE DECARBOXYLASE | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, MANGANESE (II) ION, OXALATE DECARBOXYLASE OXDC | | Authors: | Just, V.J, Stevenson, C.E.M, Bowater, L, Tanner, A, Lawson, D.M, Bornemann, S. | | Deposit date: | 2004-02-02 | | Release date: | 2004-02-19 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A Closed Conformation of Bacillus Subtilis Oxalate Decarboxylase Oxdc Provides Evidence for the True Identity of the Active Site
J.Biol.Chem., 279, 2004
|
|
6V7L
 
 | | The structure of the P212121 crystal form of canavalin at 173 K | | Descriptor: | BENZOIC ACID, Canavalin | | Authors: | McPherson, A. | | Deposit date: | 2019-12-08 | | Release date: | 2020-02-19 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Binding of benzoic acid and anions within the cupin domains of the vicilin protein canavalin from jack bean (Canavalia ensiformis): Crystal structures.
Biochem.Biophys.Res.Commun., 524, 2020
|
|
5VG3
 
 | |
5VF5
 
 | | Crystal structure of the vicilin from Solanum melongena, re-refinement | | Descriptor: | ACETATE ION, COPPER (II) ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Porebski, P.J, Wlodawer, A, Dauter, Z, Minor, W, Stanfield, R, Jaskolski, M, Pozharski, E, Weichenberger, C.X, Rupp, B. | | Deposit date: | 2017-04-06 | | Release date: | 2017-12-06 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Detect, correct, retract: How to manage incorrect structural models.
FEBS J., 285, 2018
|
|
5WPW
 
 | | Crystal structure of coconut allergen cocosin | | Descriptor: | 11S globulin isoform 1 | | Authors: | Jin, T, Zhang, Y. | | Deposit date: | 2016-11-21 | | Release date: | 2017-10-25 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.847 Å) | | Cite: | Crystal Structure of Cocosin, A Potential Food Allergen from Coconut (Cocos nucifera)
J. Agric. Food Chem., 65, 2017
|
|
5WXU
 
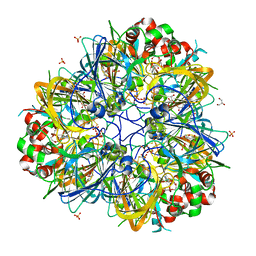 | | 11S globulin from Wrightia tinctoria reveals auxin binding site | | Descriptor: | 11S globulin, 1H-INDOL-3-YLACETIC ACID, CITRATE ANION, ... | | Authors: | Kumar, P, Kesari, P, Dhindwal, S, Kumar, P. | | Deposit date: | 2017-01-09 | | Release date: | 2018-05-23 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | A novel function for globulin in sequestering plant hormone: Crystal structure of Wrightia tinctoria 11S globulin in complex with auxin.
Sci Rep, 7, 2017
|
|
3QAC
 
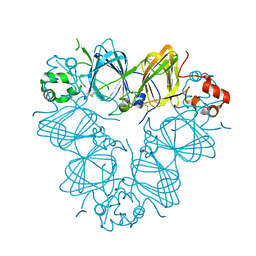 | | Structure of amaranth 11S proglobulin seed storage protein from Amaranthus hypochondriacus L. | | Descriptor: | 11S globulin seed storage protein | | Authors: | Tandang-Silvas, M.R, Carrazco-Pena, L, Barba de la Rosa, A.P, Osuna-Castro, J.A, Utsumi, S, Mikami, B, Maruyama, N. | | Deposit date: | 2011-01-10 | | Release date: | 2012-01-11 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.275 Å) | | Cite: | Structure of amaranth 11S proglobulin, a major seed storage protein from Amaranthus hypochondriacus L.
To be Published
|
|
5XTY
 
 | |
3S7I
 
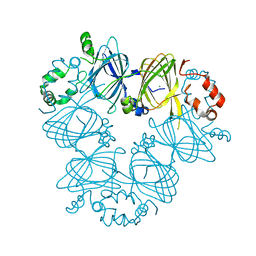 | | Crystal structure of Ara h 1 | | Descriptor: | Allergen Ara h 1, clone P41B, CHLORIDE ION | | Authors: | Chruszcz, M, Maleki, S.J, Solberg, R, Minor, W. | | Deposit date: | 2011-05-26 | | Release date: | 2011-09-21 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural and Immunologic Characterization of Ara h 1, a Major Peanut Allergen.
J.Biol.Chem., 286, 2011
|
|
3S0M
 
 | | A Structural Element that Modulates Proton-Coupled Electron Transfer in Oxalate Decarboxylase | | Descriptor: | 1,2-ETHANEDIOL, CARBONATE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Saylor, B.T, Reinhardt, L.A, Lu, Z, Shukla, M.S, Cleland, W.W, Allen, K.N, Richards, N.G.J. | | Deposit date: | 2011-05-13 | | Release date: | 2012-04-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | A structural element that facilitates proton-coupled electron transfer in oxalate decarboxylase.
Biochemistry, 51, 2012
|
|
3S7E
 
 | | Crystal structure of Ara h 1 | | Descriptor: | Allergen Ara h 1, clone P41B, CHLORIDE ION | | Authors: | Chruszcz, M, Maleki, S.J, Solberg, R, Minor, W. | | Deposit date: | 2011-05-26 | | Release date: | 2011-09-21 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.71 Å) | | Cite: | Structural and Immunologic Characterization of Ara h 1, a Major Peanut Allergen.
J.Biol.Chem., 286, 2011
|
|
3SMH
 
 | |
1CAX
 
 | | DETERMINATION OF THREE CRYSTAL STRUCTURES OF CANAVALIN BY MOLECULAR REPLACEMENT | | Descriptor: | CANAVALIN | | Authors: | Ko, T-P, Ng, J.D, Day, J, Greenwood, A, McPherson, A. | | Deposit date: | 1993-06-10 | | Release date: | 1993-10-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Determination of three crystal structures of canavalin by molecular replacement.
Acta Crystallogr.,Sect.D, 49, 1993
|
|
1CAV
 
 | |
1CAU
 
 | | DETERMINATION OF THREE CRYSTAL STRUCTURES OF CANAVALIN BY MOLECULAR REPLACEMENT | | Descriptor: | CANAVALIN | | Authors: | Ko, T-P, Ng, J.D, Day, J, Greenwood, A, McPherson, A. | | Deposit date: | 1993-07-08 | | Release date: | 1993-10-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Determination of three crystal structures of canavalin by molecular replacement.
Acta Crystallogr.,Sect.D, 49, 1993
|
|
