2EXF
 
 | |
2IHX
 
 | |
2IWJ
 
 | | SOLUTION STRUCTURE OF THE ZN COMPLEX OF HIV-2 NCP(23-49) PEPTIDE, ENCOMPASSING PROTEIN CCHC-LINKER, DISTAL CCHC ZN-BINDING MOTIF AND C- TERMINAL TAIL. | | Descriptor: | GAG-POL POLYPROTEIN, ZINC ION | | Authors: | Amodeo, P, Castiglione Morelli, M.A, Ostuni, A, Cristinziano, P, Bavoso, A. | | Deposit date: | 2006-06-30 | | Release date: | 2007-10-09 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Structural Features of the C-Terminal Zinc Finger Domain of the HIV-2 Nc Protein (Residues 23-49).
To be Published
|
|
2JZW
 
 | | How the HIV-1 nucleocapsid protein binds and destabilises the (-)primer binding site during reverse transcription | | Descriptor: | DNA (5'-D(*DGP*DTP*DCP*DCP*DCP*DTP*DGP*DTP*DTP*DCP*DGP*DGP*DGP*DC)-3'), HIV-1 nucleocapsid protein NCp7(12-55), ZINC ION | | Authors: | Bourbigot, S, Ramalanjaona, N, Salgado, G.F.J, Mely, Y, Roques, B.P, Bouaziz, S, Morellet, N. | | Deposit date: | 2008-01-21 | | Release date: | 2009-01-13 | | Last modified: | 2022-03-16 | | Method: | SOLUTION NMR | | Cite: | How the HIV-1 nucleocapsid protein binds and destabilises the (-)primer binding site during reverse transcription.
J.Mol.Biol., 383, 2008
|
|
2L45
 
 | | C-terminal zinc knuckle of the HIVNCp7 with DNA | | Descriptor: | C-TERMINAL ZINC KNUCLE OF THE HIV-NCP7, DNA (5'-D(P*TP*AP*CP*GP*CP*C)-3'), ZINC ION | | Authors: | Quintal, S, Viegas, A, Cabrita, E, Farrell, N, Erhardt, S. | | Deposit date: | 2010-10-01 | | Release date: | 2011-12-14 | | Last modified: | 2020-02-05 | | Method: | SOLUTION NMR | | Cite: | Platinated DNA affects zinc finger conformation. Interaction of a platinated single-stranded oligonucleotide and the C-terminal zinc finger of nucleocapsid protein HIVNCp7.
Biochemistry, 51, 2012
|
|
2L44
 
 | | C-terminal zinc knuckle of the HIVNCp7 | | Descriptor: | C-terminal Zinc Knucle of the HIV-NCP7, ZINC ION | | Authors: | Quintal, S.M.O, Viegas, A.J.M, Cabrita, E.J, Farrell, N.P, Erhardt, S. | | Deposit date: | 2010-10-01 | | Release date: | 2011-10-12 | | Last modified: | 2020-02-05 | | Method: | SOLUTION NMR | | Cite: | Platinated DNA affects zinc finger conformation. Interaction of a platinated single-stranded oligonucleotide and the C-terminal zinc finger of nucleocapsid protein HIVNCp7.
Biochemistry, 51, 2012
|
|
2L46
 
 | | C-terminal zinc finger of the HIVNCp7 with platinated DNA | | Descriptor: | C-TERMINAL ZINC KNUCLE OF THE HIV-NCP7, DNA (5'-D(P*TP*AP*CP*GP*CP*C)-3'), ZINC ION | | Authors: | Quintal, S, Viegas, A, Cabrita, E, Farrell, N, Erhardt, S. | | Deposit date: | 2010-10-01 | | Release date: | 2011-12-14 | | Last modified: | 2012-06-20 | | Method: | SOLUTION NMR | | Cite: | Platinated DNA affects zinc finger conformation. Interaction of a platinated single-stranded oligonucleotide and the C-terminal zinc finger of nucleocapsid protein HIVNCp7.
Biochemistry, 51, 2012
|
|
2LI8
 
 | |
2L4L
 
 | | Structural insights into the cTAR DNA recognition by the HIV-1 Nucleocapsid protein: role of sugar deoxyriboses in the binding polarity of NC | | Descriptor: | 5'-D(*CP*TP*GP*G)-3', HIV-1 nucleocapsid protein NCp7, ZINC ION | | Authors: | Bazzi, A, Zargarian, L, Chaminade, F, Boudier, C, De Rocquigny, H, Rene, B, Mely, Y, Fosse, P, Mauffret, O. | | Deposit date: | 2010-10-08 | | Release date: | 2010-12-08 | | Last modified: | 2011-08-17 | | Method: | SOLUTION NMR | | Cite: | Structural insights into the cTAR DNA recognition by the HIV-1 nucleocapsid protein: role of sugar deoxyriboses in the binding polarity of NC.
Nucleic Acids Res., 39, 2011
|
|
2M3Z
 
 | |
2MS0
 
 | |
2MPJ
 
 | | NMR structure of Xenopus RecQ4 zinc knuckle | | Descriptor: | RECQL4-helicase-like protein, ZINC ION | | Authors: | Zucchelli, C, Marino, F, Mojumdar, A, Onesti, S, Musco, G. | | Deposit date: | 2014-05-23 | | Release date: | 2015-08-05 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Structural and biochemical characterization of an RNA/DNA binding motif in the N-terminal domain of RecQ4 helicases
Sci Rep, 6, 2016
|
|
2MS1
 
 | |
2MQV
 
 | |
3NYB
 
 | |
8C9M
 
 | | HERV-K Gag immature lattice | | Descriptor: | Gag protein | | Authors: | Krebs, A.-S, Liu, H.-F, Zhou, Y, Rey, J.S, Levintov, L, Perilla, J.R, Bartesaghi, A, Zhang, P. | | Deposit date: | 2023-01-23 | | Release date: | 2023-02-01 | | Last modified: | 2023-07-05 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Molecular architecture and conservation of an immature human endogenous retrovirus.
Biorxiv, 2023
|
|
5A9E
 
 | | Cryo-electron tomography and subtomogram averaging of Rous-Sarcoma- Virus deltaMBD virus-like particles | | Descriptor: | DELTAMBD GAG PROTEIN | | Authors: | Schur, F.K.M, Dick, R.A, Hagen, W.J.H, Vogt, V.M, Briggs, J.A.G. | | Deposit date: | 2015-07-21 | | Release date: | 2015-08-12 | | Last modified: | 2017-08-02 | | Method: | ELECTRON MICROSCOPY (7.7 Å) | | Cite: | The Structure of Immature-Like Rous Sarcoma Virus Gag Particles Reveals a Structural Role for the P10 Domain in Assembly.
J.Virol., 89, 2015
|
|
5O2U
 
 | | Llama VHH in complex with p24 | | Descriptor: | Capsid protein p24, VHH 59H10 | | Authors: | Caillat, C, Verrips, T, Weissenhorn, W. | | Deposit date: | 2017-05-22 | | Release date: | 2017-06-21 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.76 Å) | | Cite: | Unravelling the Molecular Basis of High Affinity Nanobodies against HIV p24: In Vitro Functional, Structural, and in Silico Insights.
ACS Infect Dis, 3, 2017
|
|
6DNH
 
 | | Cryo-EM structure of human CPSF-160-WDR33-CPSF-30-PAS RNA complex at 3.4 A resolution | | Descriptor: | Cleavage and polyadenylation specificity factor subunit 1, Cleavage and polyadenylation specificity factor subunit 4, RNA (5'-R(P*AP*AP*UP*AP*AP*AP*C)-3'), ... | | Authors: | Sun, Y, Zhang, Y, Hamilton, K, Walz, T, Tong, L. | | Deposit date: | 2018-06-06 | | Release date: | 2018-06-27 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Molecular basis for the recognition of the human AAUAAA polyadenylation signal.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
8E3I
 
 | | CRYO-EM STRUCTURE OF the human MPSF IN COMPLEX WITH THE AUUAAA poly(A) signal | | Descriptor: | Cleavage and polyadenylation specificity factor subunit 1, Cleavage and polyadenylation specificity factor subunit 4, RNA (5'-R(P*CP*AP*UP*UP*AP*AP*AP*CP*AP*AP*C)-3'), ... | | Authors: | Gutierrez, P.A, Wei, J, Sun, Y, Tong, L. | | Deposit date: | 2022-08-17 | | Release date: | 2023-01-18 | | Method: | ELECTRON MICROSCOPY (2.53 Å) | | Cite: | Molecular basis for the recognition of the AUUAAA polyadenylation signal by mPSF.
Rna, 28, 2022
|
|
8E3Q
 
 | | CRYO-EM STRUCTURE OF the human MPSF | | Descriptor: | Cleavage and polyadenylation specificity factor subunit 1, Cleavage and polyadenylation specificity factor subunit 4, ZINC ION, ... | | Authors: | Gutierrez, P.A, Wei, J, Sun, Y, Tong, L. | | Deposit date: | 2022-08-17 | | Release date: | 2023-01-18 | | Method: | ELECTRON MICROSCOPY (2.68 Å) | | Cite: | Molecular basis for the recognition of the AUUAAA polyadenylation signal by mPSF.
Rna, 28, 2022
|
|
6URG
 
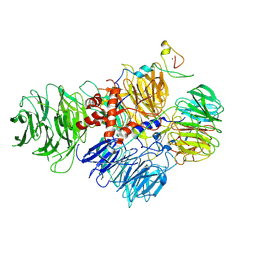 | | Cryo-EM structure of human CPSF160-WDR33-CPSF30-CPSF100 PIM complex | | Descriptor: | Cleavage and polyadenylation specificity factor subunit 1, Cleavage and polyadenylation specificity factor subunit 2, Cleavage and polyadenylation specificity factor subunit 4, ... | | Authors: | Sun, Y, Zhang, Y, Walz, T, Tong, L. | | Deposit date: | 2019-10-23 | | Release date: | 2019-11-27 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural Insights into the Human Pre-mRNA 3'-End Processing Machinery.
Mol.Cell, 77, 2020
|
|
6URO
 
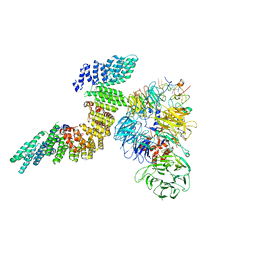 | | Cryo-EM structure of human CPSF160-WDR33-CPSF30-PAS RNA-CstF77 complex | | Descriptor: | Cleavage and polyadenylation specificity factor subunit 1, Cleavage and polyadenylation specificity factor subunit 4, Cleavage stimulation factor subunit 3, ... | | Authors: | Sun, Y, Zhang, Y, Walz, T, Tong, L. | | Deposit date: | 2019-10-23 | | Release date: | 2019-11-27 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structural Insights into the Human Pre-mRNA 3'-End Processing Machinery.
Mol.Cell, 77, 2020
|
|
8PUG
 
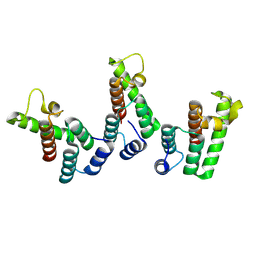 | | Structure of the immature HTLV-1 CA lattice from full-length Gag VLPs: CA-NTD refinement | | Descriptor: | Gag polyprotein | | Authors: | Obr, M, Percipalle, M, Chernikova, D, Yang, H, Thader, A, Pinke, G, Porley, D, Mansky, L.M, Dick, R.A, Schur, F.K.M. | | Deposit date: | 2023-07-17 | | Release date: | 2023-08-23 | | Method: | ELECTRON MICROSCOPY (5.9 Å) | | Cite: | Unconventional stabilization of the human T-cell leukemia virus type 1 immature Gag lattice.
Biorxiv, 2023
|
|
8PUH
 
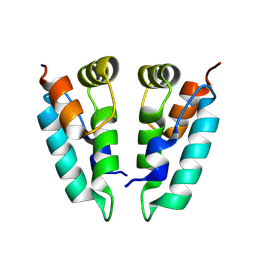 | | Structure of the immature HTLV-1 CA lattice from full-length Gag VLPs: CA-CTD refinement | | Descriptor: | Gag polyprotein | | Authors: | Obr, M, Percipalle, M, Chernikova, D, Yang, H, Thader, A, Pinke, G, Porley, D, Mansky, L.M, Dick, R.A, Schur, F.K.M. | | Deposit date: | 2023-07-17 | | Release date: | 2023-08-23 | | Method: | ELECTRON MICROSCOPY (6.2 Å) | | Cite: | Unconventional stabilization of the human T-cell leukemia virus type 1 immature Gag lattice.
Biorxiv, 2023
|
|
