5T1I
 
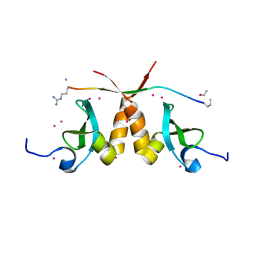 | | CBX3 chromo shadow domain in complex with histone H3 peptide | | Descriptor: | Chromobox protein homolog 3, Histone H3.1, UNKNOWN ATOM OR ION | | Authors: | Liu, Y, Tempel, W, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2016-08-19 | | Release date: | 2016-09-14 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Peptide recognition by heterochromatin protein 1 (HP1) chromoshadow domains revisited: Plasticity in the pseudosymmetric histone binding site of human HP1.
J. Biol. Chem., 292, 2017
|
|
7OS4
 
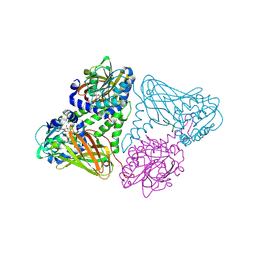 | | Crystal structure of mouse CARM1 in complex with histone H3_13-31 K18 | | Descriptor: | (2~{R},3~{R},4~{S},5~{R})-2-(6-aminopurin-9-yl)-5-[(~{E})-prop-1-enyl]oxolane-3,4-diol, Histone H3.1, Histone-arginine methyltransferase CARM1 | | Authors: | Marechal, N, Cura, V, Troffer-Charlier, N, Bonnefond, L, Cavarelli, J. | | Deposit date: | 2021-06-07 | | Release date: | 2021-10-27 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.54 Å) | | Cite: | Structural Studies Provide New Insights into the Role of Lysine Acetylation on Substrate Recognition by CARM1 and Inform the Design of Potent Peptidomimetic Inhibitors.
Chembiochem, 22, 2021
|
|
7OKP
 
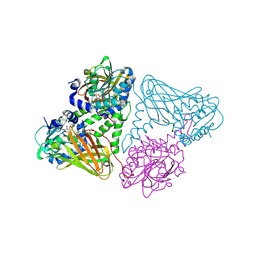 | | Crystal structure of mouse CARM1 in complex with histone H3_13-22 K18 acetylated | | Descriptor: | (2~{R},3~{R},4~{S},5~{R})-2-(6-aminopurin-9-yl)-5-[(~{E})-prop-1-enyl]oxolane-3,4-diol, Histone H3.3, Histone-arginine methyltransferase CARM1, ... | | Authors: | Marechal, N, Cura, V, Troffer-Charlier, N, Bonnefond, L, Cavarelli, J. | | Deposit date: | 2021-05-18 | | Release date: | 2021-10-27 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural Studies Provide New Insights into the Role of Lysine Acetylation on Substrate Recognition by CARM1 and Inform the Design of Potent Peptidomimetic Inhibitors.
Chembiochem, 22, 2021
|
|
7K7G
 
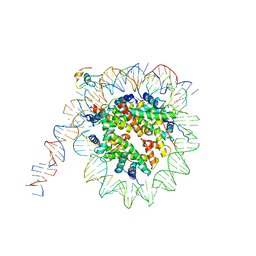 | | nucleosome and Gal4 complex | | Descriptor: | Centromere DNA-binding protein complex CBF3 subunit B, DNA (147-MER), Histone H2A.1, ... | | Authors: | Ruifang, G, Yawen, B. | | Deposit date: | 2020-09-22 | | Release date: | 2021-03-31 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Structural and dynamic mechanisms of CBF3-guided centromeric nucleosome formation.
Nat Commun, 12, 2021
|
|
2HUE
 
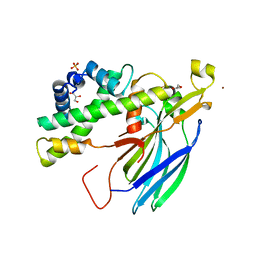 | | Structure of the H3-H4 chaperone Asf1 bound to histones H3 and H4 | | Descriptor: | Anti-silencing protein 1, GLYCEROL, Histone H3, ... | | Authors: | English, C.M, Churchill, M.E.A, Tyler, J.K. | | Deposit date: | 2006-07-26 | | Release date: | 2006-11-21 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural basis for the histone chaperone activity of asf1.
Cell(Cambridge,Mass.), 127, 2006
|
|
2LH0
 
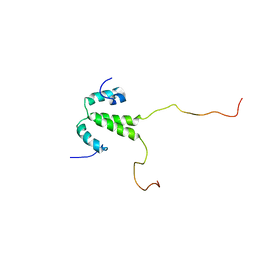 | |
6YN1
 
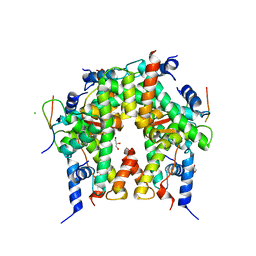 | | Crystal structure of histone chaperone APLF acidic domain bound to the histone H2A-H2B-H3-H4 octamer | | Descriptor: | Aprataxin and PNK-like factor, CHLORIDE ION, GLYCEROL, ... | | Authors: | Corbeski, I, Guo, X, Van Ingen, H, Sixma, T.K. | | Deposit date: | 2020-04-10 | | Release date: | 2021-11-17 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Chaperoning of the histone octamer by the acidic domain of DNA repair factor APLF.
Sci Adv, 8, 2022
|
|
7CJ0
 
 | |
7CIZ
 
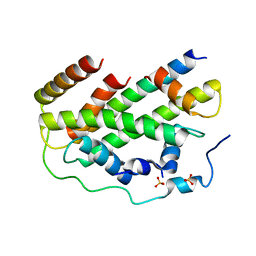 | |
7Y5V
 
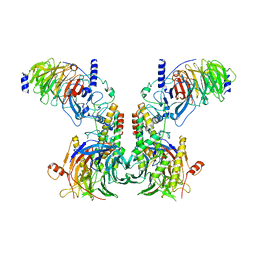 | | Cryo-EM structure of the dimeric human CAF1LC-H3-H4 complex | | Descriptor: | Chromatin assembly factor 1 subunit A, Chromatin assembly factor 1 subunit B, Histone H3.1, ... | | Authors: | Liu, C.P, Yu, C, Yu, Z.Y, Xu, R.M. | | Deposit date: | 2022-06-17 | | Release date: | 2023-08-16 | | Last modified: | 2023-09-06 | | Method: | ELECTRON MICROSCOPY (6.1 Å) | | Cite: | Structural insights into histone binding and nucleosome assembly by chromatin assembly factor-1.
Science, 381, 2023
|
|
7Y5U
 
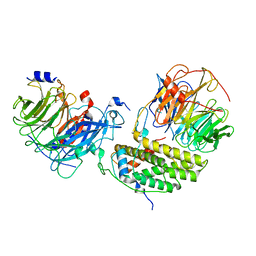 | | Cryo-EM structure of the monomeric human CAF1LC-H3-H4 complex | | Descriptor: | Chromatin assembly factor 1 subunit A, Chromatin assembly factor 1 subunit B, Histone H3.1, ... | | Authors: | Liu, C.P, Yu, Z.Y, Yu, C, Xu, R.M. | | Deposit date: | 2022-06-17 | | Release date: | 2023-08-16 | | Last modified: | 2023-09-06 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structural insights into histone binding and nucleosome assembly by chromatin assembly factor-1.
Science, 381, 2023
|
|
8P88
 
 | |
8P89
 
 | |
5KDM
 
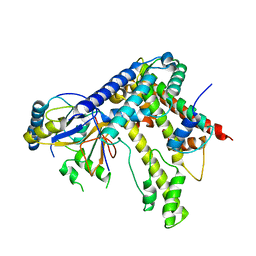 | |
5Z23
 
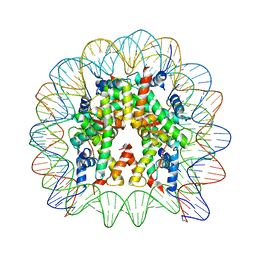 | | Crystal structure of the nucleosome containing a chimeric histone H3/CENP-A CATD | | Descriptor: | DNA (146-MER), Histone H2A type 1-B/E, Histone H2B type 1-J, ... | | Authors: | Arimura, Y, Tachiwana, H, Takagi, H. | | Deposit date: | 2017-12-28 | | Release date: | 2019-02-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.73 Å) | | Cite: | The CENP-A centromere targeting domain facilitates H4K20 monomethylation in the nucleosome by structural polymorphism.
Nat Commun, 10, 2019
|
|
5ZBX
 
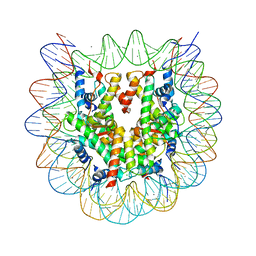 | | The crystal structure of the nucleosome containing histone H3.1 CATD(V76Q, K77D) | | Descriptor: | CHLORIDE ION, DNA (146-MER), Histone H2A type 1-B/E, ... | | Authors: | Arimura, Y, Takagi, H, Kurumizaka, H. | | Deposit date: | 2018-02-13 | | Release date: | 2019-02-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.58 Å) | | Cite: | The CENP-A centromere targeting domain facilitates H4K20 monomethylation in the nucleosome by structural polymorphism.
Nat Commun, 10, 2019
|
|
7KF0
 
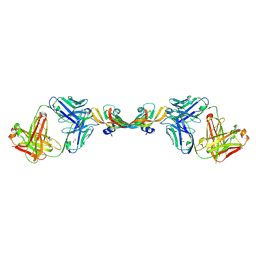 | |
7KF1
 
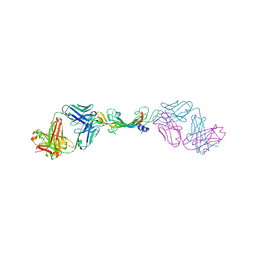 | |
7KEZ
 
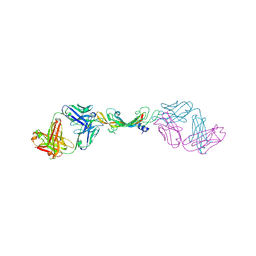 | |
4QEO
 
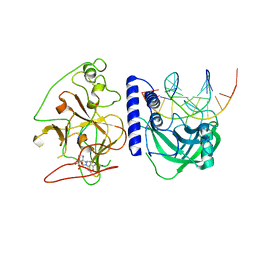 | | crystal structure of KRYPTONITE in complex with mCHH DNA, H3(1-15) peptide and SAH | | Descriptor: | DNA 5'-ACTGATGAGTACCAT-3', DNA 5'-GGTACT(5CM)ATCAGTAT-3', Histone H3, ... | | Authors: | Du, J, Li, S, Patel, D.J. | | Deposit date: | 2014-05-17 | | Release date: | 2014-07-30 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Mechanism of DNA Methylation-Directed Histone Methylation by KRYPTONITE.
Mol.Cell, 55, 2014
|
|
2F6J
 
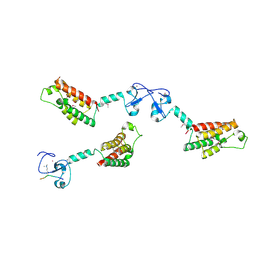 | |
2FSA
 
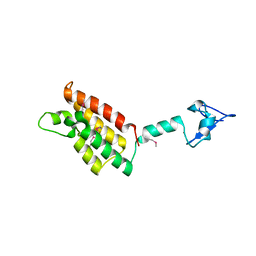 | |
6WW3
 
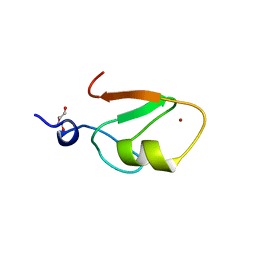 | | Crystal structure of HERC2 ZZ domain in complex with SUMO1 tail | | Descriptor: | DI(HYDROXYETHYL)ETHER, GLYCEROL, SUMO1 linked HERC2 ZZ domain (Small ubiquitin-related modifier 1,E3 ubiquitin-protein ligase HERC2), ... | | Authors: | Liu, J, Vann, K.R, Kutateladze, T.G. | | Deposit date: | 2020-05-07 | | Release date: | 2020-08-05 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.096 Å) | | Cite: | Structural Insight into Binding of the ZZ Domain of HERC2 to Histone H3 and SUMO1.
Structure, 28, 2020
|
|
7D86
 
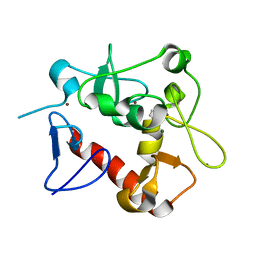 | | Crystal Structure of zebrafishPHF14-PZP | | Descriptor: | 1,2-ETHANEDIOL, MAGNESIUM ION, PHD finger protein 14, ... | | Authors: | Li, H, Zheng, S. | | Deposit date: | 2020-10-07 | | Release date: | 2021-07-28 | | Last modified: | 2022-02-09 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Molecular basis for bipartite recognition of histone H3 by the PZP domain of PHF14.
Nucleic Acids Res., 49, 2021
|
|
3A1B
 
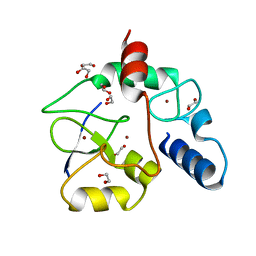 | | Crystal structure of the DNMT3A ADD domain in complex with histone H3 | | Descriptor: | 1,2-ETHANEDIOL, DNA (cytosine-5)-methyltransferase 3A, Histone H3.1, ... | | Authors: | Otani, J, Arita, K, Ariyoshi, M, Shirakawa, M. | | Deposit date: | 2009-03-28 | | Release date: | 2009-11-10 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.292 Å) | | Cite: | Structural basis for recognition of H3K4 methylation status by the DNA methyltransferase 3A ATRX-DNMT3-DNMT3L domain
Embo Rep., 10, 2009
|
|
