8SKE
 
 | |
8SKT
 
 | |
8SKX
 
 | |
8SL5
 
 | | [2T13] Self-assembling left-handed two-turn tensegrity triangle with 13 interjunction base pairs and R3 symmetry | | Descriptor: | DNA (5'-D(*GP*AP*GP*CP*CP*TP*GP*AP*CP*TP*AP*CP*A)-3'), DNA (5'-D(*TP*CP*TP*GP*TP*GP*GP*C)-3'), DNA (5'-D(P*CP*GP*TP*GP*GP*AP*CP*A)-3'), ... | | Authors: | Vecchioni, S, Janowski, J, Sha, R, Ohayon, Y.P. | | Deposit date: | 2023-04-21 | | Release date: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (6.55 Å) | | Cite: | Programming Tertiary Chirality in Self-Assembling 3D DNA Crystals
To Be Published
|
|
8SVL
 
 | |
8SVM
 
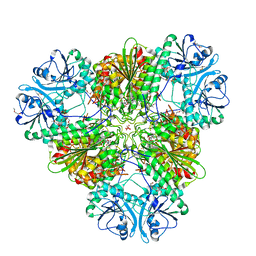 | | Plasmodium falciparum M17 aminopeptidase bound to MMV1557817 | | Descriptor: | CARBONATE ION, Leucine aminopeptidase, N-[(1R)-2-(hydroxyamino)-2-oxo-1-(3',4',5'-trifluoro[1,1'-biphenyl]-4-yl)ethyl]-3,3-dimethylbutanamide, ... | | Authors: | McGowan, S, Drinkwater, N. | | Deposit date: | 2023-05-17 | | Release date: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Characterisation of a novel antimalarial agent targeting haemaglobin digestion that shows cross-species reactivity and excellent in vivo properties.
Mbio, 2024
|
|
8SW9
 
 | | Plasmodium falciparum M17 (A460S) mutant | | Descriptor: | CARBONATE ION, Leucine aminopeptidase, PENTAETHYLENE GLYCOL, ... | | Authors: | McGowan, S, Suraweera, C, Drinkwater, N. | | Deposit date: | 2023-05-17 | | Release date: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Characterisation of a novel antimalarial agent targeting haemaglobin digestion that shows cross-species reactivity and excellent in vivo properties.
Mbio, 2024
|
|
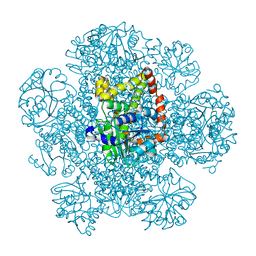
8SWM
 
 | |
8SXD
 
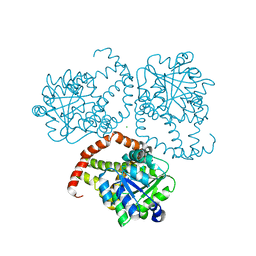 | |
8T6C
 
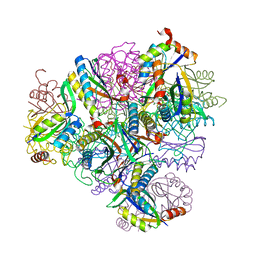 | | Crystal structure of T33-18.2: Deep-learning sequence design of co-assembling tetrahedron protein nanoparticles | | Descriptor: | T33-18.2 : A, T33-18.2 : B | | Authors: | Bera, A.K, de Haas, R.J, Kang, A, Sankaran, B, King, N.P. | | Deposit date: | 2023-06-15 | | Release date: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Rapid and automated design of two-component protein nanomaterials using ProteinMPNN.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8T6E
 
 | | Crystal structure of T33-28.3: Deep-learning sequence design of co-assembling tetrahedron protein nanoparticles | | Descriptor: | T33-28.3: A, T33-28.3: B | | Authors: | Bera, A.K, de Haas, R.J, Kang, A, Sankaran, B, King, N.P. | | Deposit date: | 2023-06-15 | | Release date: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Rapid and automated design of two-component protein nanomaterials using ProteinMPNN.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8T6N
 
 | | Crystal structure of T33-27.1: Deep-learning sequence design of co-assembling tetrahedron protein nanoparticles | | Descriptor: | T33-27.1 : A, T33-27.1 : B | | Authors: | Bera, A.K, de Haas, R.J, Kang, A, Sankaran, B, King, N.P. | | Deposit date: | 2023-06-16 | | Release date: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (3.63 Å) | | Cite: | Rapid and automated design of two-component protein nanomaterials using ProteinMPNN.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8TJY
 
 | | Structure of Gabija AB complex | | Descriptor: | Endonuclease GajA, Gabija protein GajB | | Authors: | Shen, Z.F, Yang, X.Y, Fu, T.M. | | Deposit date: | 2023-07-24 | | Release date: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (2.79 Å) | | Cite: | Molecular basis of Gabija anti-phage supramolecular assemblies
Nat.Struct.Mol.Biol., 2024
|
|
8TK0
 
 | |
8TK1
 
 | | Structure of Gabija AB complex 1 | | Descriptor: | Endonuclease GajA, Gabija protein GajB | | Authors: | Shen, Z.F, Yang, X.Y, Fu, T.M. | | Deposit date: | 2023-07-25 | | Release date: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (2.98 Å) | | Cite: | Molecular basis of Gabija anti-phage supramolecular assemblies
Nat.Struct.Mol.Biol., 2024
|
|
8TN1
 
 | |
8TN6
 
 | |
8TNB
 
 | |
8TNC
 
 | |
8TND
 
 | |
8TQD
 
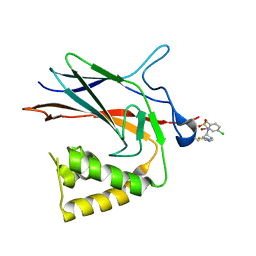 | | NF-Kappa-B1 Bound with a Covalent Inhibitor | | Descriptor: | 1-(2-bromo-4-chlorophenyl)-N-{(3S)-1-[(E)-iminomethyl]pyrrolidin-3-yl}methanesulfonamide, Nuclear factor NF-kappa-B p105 subunit | | Authors: | Hilbert, B.J. | | Deposit date: | 2023-08-07 | | Release date: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | DrugMap: A quantitative pan-cancer analysis of cysteine ligandability
To Be Published
|
|
8TQG
 
 | |
8TQI
 
 | |
8TQK
 
 | |
8TQV
 
 | |
