2DUE
 
 | |
2QT2
 
 | | Cyclized-Dehydrated Intermediate of GFP Variant Q183E in Chromophore Maturation | | Descriptor: | 1,2-ETHANEDIOL, Green fluorescent protein | | Authors: | Wood, T.I, Barondeau, D.P, Hitomi, C, Tainer, J.A, Getzoff, E.D. | | Deposit date: | 2007-07-31 | | Release date: | 2009-04-21 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.31 Å) | | Cite: | Kinetically Isolated Reaction Intermediates Provide Structural Characterization of the Green Fluorescence Protein Fluorophore Biosynthesis Pathway
To be Published
|
|
1RMP
 
 | | Probing the Role of Tryptophans in Aequorea Victoria Green Fluorescent Proteins with an Expanded Genetic Code | | Descriptor: | SIGF1-GFP fusion protein | | Authors: | Budisa, N, Pal, P.P, Alefelder, S, Birle, P, Krywcun, T, Rubini, M, Wenger, W, Bae, J.H, Steiner, T. | | Deposit date: | 2003-11-28 | | Release date: | 2004-06-08 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Probing the role of tryptophans in Aequorea victoria green fluorescent proteins with an expanded genetic code
Biol.Chem., 385, 2004
|
|
1RMM
 
 | | Probing the Role of Tryptophans in Aequorea Victoria Green Fluorescent Proteins with an Expanded Genetic Code | | Descriptor: | SIGF1-GFP fusion protein | | Authors: | Budisa, N, Pal, P.P, Alefelder, S, Birle, P, Krywcun, T, Rubini, M, Wenger, W, Bae, J.H, Steiner, T. | | Deposit date: | 2003-11-28 | | Release date: | 2004-06-08 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Probing the role of tryptophans in Aequorea victoria green fluorescent proteins with an expanded genetic code
Biol.Chem., 385, 2004
|
|
1RMO
 
 | | Probing the Role of Tryptophans in Aequorea Victoria Green Fluorescent Proteins with an Expanded Genetic Code | | Descriptor: | wunen-nonfunctional GFP fusion protein | | Authors: | Budisa, N, Pal, P.P, Alefelder, S, Birle, P, Krywcun, T, Rubini, M, Wenger, W, Bae, J.H, Steiner, T. | | Deposit date: | 2003-11-28 | | Release date: | 2004-06-08 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Probing the role of tryptophans in Aequorea victoria green fluorescent proteins with an expanded genetic code
Biol.Chem., 385, 2004
|
|
1RRX
 
 | | Crystallographic Evidence for Isomeric Chromophores in 3-Fluorotyrosyl-Green Fluorescent Protein | | Descriptor: | SIGF1-GFP fusion protein | | Authors: | Bae, J.H, Paramita Pal, P, Moroder, L, Huber, R, Budisa, N. | | Deposit date: | 2003-12-09 | | Release date: | 2004-06-08 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystallographic Evidence for Isomeric Chromophores in 3-Fluorotyrosyl-Green Fluorescent Protein.
Chembiochem, 5, 2004
|
|
7SAH
 
 | |
7SAI
 
 | | Crystal Structure of Lag30 Nanobody bound to eGFP | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, GLYCEROL, Green fluorescent protein, ... | | Authors: | Cong, A.T.Q, Schellenberg, M.J. | | Deposit date: | 2021-09-22 | | Release date: | 2022-09-14 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | High-efficiency recombinant protein purification using mCherry and YFP nanobody affinity matrices.
Protein Sci., 31, 2022
|
|
1JC0
 
 | | CRYSTAL STRUCTURE ANALYSIS OF A REDOX-SENSITIVE GREEN FLUORESCENT PROTEIN VARIANT IN A REDUCED FORM | | Descriptor: | GREEN FLUORESCENT PROTEIN | | Authors: | Hanson, G.T, Aggeler, R, Oglesbee, D, Cannon, M, Capaldi, R.A, Tsien, R.Y, Remington, S.J. | | Deposit date: | 2001-06-07 | | Release date: | 2003-09-09 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Investigating mitochondrial redox potential with redox-sensitive green fluorescent protein indicators.
J.Biol.Chem., 279, 2004
|
|
1JC1
 
 | | CRYSTAL STRUCTURE ANALYSIS OF A REDOX-SENSITIVE GREEN FLUORESCENT PROTEIN VARIANT IN A OXIDIZED FORM | | Descriptor: | GREEN FLUORESCENT PROTEIN | | Authors: | Hanson, G.T, Aggeler, R, Oglesbee, D, Cannon, M, Capaldi, R.A, Tsien, R.Y, Remington, S.J. | | Deposit date: | 2001-06-07 | | Release date: | 2003-09-09 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Investigating mitochondrial redox potential with redox-sensitive green fluorescent protein indicators.
J.Biol.Chem., 279, 2004
|
|
2HJO
 
 | | Crystal structure of V224H design intermediate for GFP metal ion reporter | | Descriptor: | 1,2-ETHANEDIOL, Green Fluorescent Protein, MAGNESIUM ION | | Authors: | Barondeau, D.P, Tubbs, J.L, Tainer, J.A, Getzoff, E.D. | | Deposit date: | 2006-06-30 | | Release date: | 2008-04-08 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Iterative Structure-Based Design of a Green
Fluorescent Protein Metal Ion Reporter
To be Published
|
|
2HQZ
 
 | | Crystal structure of L42H design intermediate for GFP metal ion reporter | | Descriptor: | 1,2-ETHANEDIOL, Green fluorescent protein, MAGNESIUM ION | | Authors: | Barondeau, D.P, Tubbs, J.L, Tainer, J.A, Getzoff, E.D. | | Deposit date: | 2006-07-19 | | Release date: | 2008-04-08 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Iterative Structure-Based Design of a Green Fluorescent Protein Metal Ion Reporter
To be Published
|
|
2A50
 
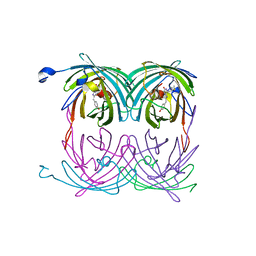 | | fluorescent protein asFP595, wt, off-state | | Descriptor: | GFP-like non-fluorescent chromoprotein FP595 chain 1, GFP-like non-fluorescent chromoprotein FP595 chain 2 | | Authors: | Andresen, M, Wahl, M.C, Stiel, A.C, Graeter, F, Schaefer, L, Trowitzsch, S, Weber, G, Eggeling, C, Grubmueller, H, Hell, S.W, Jakobs, S. | | Deposit date: | 2005-06-30 | | Release date: | 2005-08-16 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structure and mechanism of the reversible photoswitch of a fluorescent protein
Proc.Natl.Acad.Sci.Usa, 102, 2005
|
|
6IR7
 
 | | Green fluorescent protein variant GFPuv with the modification to 6-hydroxynorleucine at the C-terminus | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 6-HYDROXY-L-NORLEUCINE, Green fluorescent protein, ... | | Authors: | Nakatani, T, Yasui, N, Yamashita, A. | | Deposit date: | 2018-11-12 | | Release date: | 2019-04-03 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.277 Å) | | Cite: | Specific modification at the C-terminal lysine residue of the green fluorescent protein variant, GFPuv, expressed in Escherichia coli.
Sci Rep, 9, 2019
|
|
6IR6
 
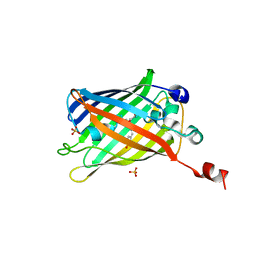 | | Green fluorescent protein variant GFPuv with the native lysine residue at the C-terminus | | Descriptor: | Green fluorescent protein, SULFATE ION | | Authors: | Nakatani, T, Yasui, N, Yamashita, A. | | Deposit date: | 2018-11-12 | | Release date: | 2019-04-03 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.642 Å) | | Cite: | Specific modification at the C-terminal lysine residue of the green fluorescent protein variant, GFPuv, expressed in Escherichia coli.
Sci Rep, 9, 2019
|
|
8DTA
 
 | |
7PD0
 
 | | Functional and structural characterization of redox sensitive superfolder green fluorescent protein and variants | | Descriptor: | DI(HYDROXYETHYL)ETHER, GLYCEROL, Green fluorescent protein, ... | | Authors: | Fritz-Wolf, K, Heimsch, K.C, Schuh, A.K, Becker, K. | | Deposit date: | 2021-08-04 | | Release date: | 2022-02-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure and Function of Redox-Sensitive Superfolder Green Fluorescent Protein Variant.
Antioxid.Redox Signal., 37, 2022
|
|
7PCA
 
 | | Functional and structural characterization of redox sensitive superfolder green fluorescent protein and variants | | Descriptor: | ETHANOL, FORMAMIDE, GLYCEROL, ... | | Authors: | Fritz-Wolf, K, Heimsch, K.C, Schuh, A.K, Becker, K. | | Deposit date: | 2021-08-03 | | Release date: | 2022-02-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Structure and Function of Redox-Sensitive Superfolder Green Fluorescent Protein Variant.
Antioxid.Redox Signal., 37, 2022
|
|
7PCZ
 
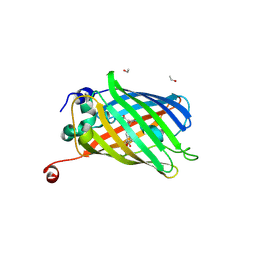 | | Functional and structural characterization of redox sensitive superfolder green fluorescent protein and variants | | Descriptor: | ETHANOL, GLYCEROL, Green fluorescent protein | | Authors: | Fritz-Wolf, K, Heimsch, K.C, Schuh, A.K, Becker, K. | | Deposit date: | 2021-08-04 | | Release date: | 2022-02-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Structure and Function of Redox-Sensitive Superfolder Green Fluorescent Protein Variant.
Antioxid.Redox Signal., 37, 2022
|
|
5DTX
 
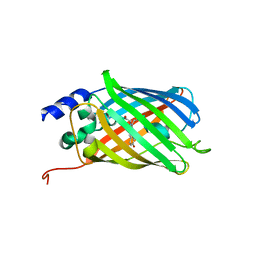 | | Crystal structure of rsEGFP2 in the fluorescent on-state | | Descriptor: | Green fluorescent protein | | Authors: | Adam, V, Martins, A. | | Deposit date: | 2015-09-18 | | Release date: | 2016-01-20 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Rational design of ultrastable and reversibly photoswitchable fluorescent proteins for super-resolution imaging of the bacterial periplasm.
Sci Rep, 6, 2016
|
|
5DTZ
 
 | |
5DTY
 
 | |
5DU0
 
 | |
1EMA
 
 | |
4GF6
 
 | | crystal structure of GFP with cuprum bound at the Incorporated metal Chelating Amino Acid PYZ151 | | Descriptor: | CALCIUM ION, COPPER (II) ION, green fluorescent protein | | Authors: | Dong, J, Liu, X, Li, J, Wang, J, Gong, W. | | Deposit date: | 2012-08-03 | | Release date: | 2012-08-29 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Genetic incorporation of a metal-chelating amino Acid as a probe for protein electron transfer.
Angew.Chem.Int.Ed.Engl., 51, 2012
|
|
