8XVZ
 
 | |
8XVY
 
 | |
8XVX
 
 | |
8XVF
 
 | |
8XV9
 
 | | Fedratinib-bound human SLC19A3 | | Descriptor: | N-tert-butyl-3-{[5-methyl-2-({4-[2-(pyrrolidin-1-yl)ethoxy]phenyl}amino)pyrimidin-4-yl]amino}benzenesulfonamide, Soluble cytochrome b562,Thiamine transporter 2 | | Authors: | Dang, Y, Wang, G.P, Zhang, Z. | | Deposit date: | 2024-01-14 | | Release date: | 2024-03-27 | | Last modified: | 2024-04-03 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Substrate and drug recognition mechanisms of SLC19A3.
Cell Res., 2024
|
|
8XV5
 
 | | Pyridoxamine-bound human SLC19A3 | | Descriptor: | 4-(AMINOMETHYL)-5-(HYDROXYMETHYL)-2-METHYLPYRIDIN-3-OL, Soluble cytochrome b562,Thiamine transporter 2 | | Authors: | Dang, Y, Wang, G.P, Zhang, Z. | | Deposit date: | 2024-01-14 | | Release date: | 2024-03-27 | | Last modified: | 2024-04-03 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Substrate and drug recognition mechanisms of SLC19A3.
Cell Res., 2024
|
|
8XV2
 
 | | Thiamine-bound human SLC19A3 | | Descriptor: | 3-(4-AMINO-2-METHYL-PYRIMIDIN-5-YLMETHYL)-5-(2-HYDROXY-ETHYL)-4-METHYL-THIAZOL-3-IUM, Soluble cytochrome b562,Thiamine transporter 2 | | Authors: | Dang, Y, Wang, G.P, Zhang, Z. | | Deposit date: | 2024-01-14 | | Release date: | 2024-03-27 | | Last modified: | 2024-04-03 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Substrate and drug recognition mechanisms of SLC19A3.
Cell Res., 2024
|
|
8XU5
 
 | |
8XU4
 
 | | The Crystal Structure of MAPK2 from Biortus. | | Descriptor: | MALONIC ACID, MAP kinase-activated protein kinase 2 | | Authors: | Wang, F, Cheng, W, Yuan, Z, Qi, J, Shen, Z. | | Deposit date: | 2024-01-12 | | Release date: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | The Crystal Structure of MAPK2 from Biortus.
To Be Published
|
|
8XTH
 
 | |
8XS5
 
 | |
8XS4
 
 | |
8XS0
 
 | |
8XRY
 
 | |
8XRX
 
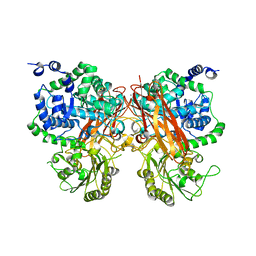 | | The crystal structure of a GH3 enzyme CcBgl3B with glucose and gentiobiose | | Descriptor: | CALCIUM ION, GH3 enzyme CcBgl3B, alpha-D-glucopyranose, ... | | Authors: | Su, J.Y. | | Deposit date: | 2024-01-08 | | Release date: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A trapped covalent intermediate as a key catalytic element in the hydrolysis of a GH3 beta-glucosidase: An X-ray crystallographic and biochemical study.
Int.J.Biol.Macromol., 265, 2024
|
|
8XRV
 
 | | The crystal structure of a GH3 enzyme CcBgl3B with glucose | | Descriptor: | CALCIUM ION, GH3 enzyme CcBgl3B, beta-D-glucopyranose | | Authors: | Su, J.Y. | | Deposit date: | 2024-01-08 | | Release date: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | A trapped covalent intermediate as a key catalytic element in the hydrolysis of a GH3 beta-glucosidase: An X-ray crystallographic and biochemical study.
Int.J.Biol.Macromol., 265, 2024
|
|
8XRU
 
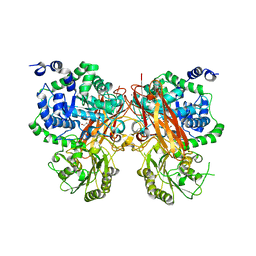 | | The crystal structure of a GH3 enzyme CcBgl3B with glycerol | | Descriptor: | CALCIUM ION, GH3 enzyme CcBgl3B, GLYCEROL | | Authors: | Su, J.Y. | | Deposit date: | 2024-01-08 | | Release date: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | A trapped covalent intermediate as a key catalytic element in the hydrolysis of a GH3 beta-glucosidase: An X-ray crystallographic and biochemical study.
Int.J.Biol.Macromol., 265, 2024
|
|
8XRT
 
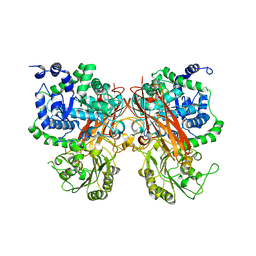 | | The crystal structure of a GH3 enzyme CcBgl3B | | Descriptor: | CALCIUM ION, GH3 enzyme CcBgl3B | | Authors: | Su, J.Y. | | Deposit date: | 2024-01-08 | | Release date: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | A trapped covalent intermediate as a key catalytic element in the hydrolysis of a GH3 beta-glucosidase: An X-ray crystallographic and biochemical study.
Int.J.Biol.Macromol., 265, 2024
|
|
8XR4
 
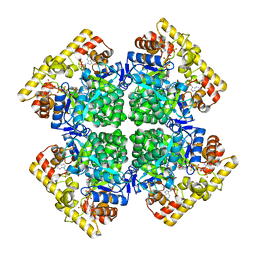 | | Crystal structure of AKRtyl-NADP(H) complex | | Descriptor: | Aldo/keto reductase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Lin, S, Dai, S, Xiao, Z. | | Deposit date: | 2024-01-06 | | Release date: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | A three-level regulatory mechanism of the aldo-keto reductase subfamily AKR12D
Nat Commun, 15, 2024
|
|
8XR3
 
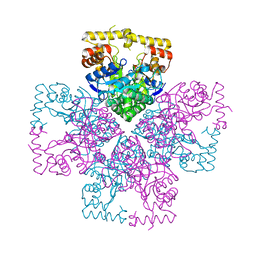 | | Crystal structure of AKRtyl-apo2 | | Descriptor: | Aldo/keto reductase | | Authors: | Lin, S, Dai, S, Xiao, Z. | | Deposit date: | 2024-01-06 | | Release date: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | A three-level regulatory mechanism of the aldo-keto reductase subfamily AKR12D
Nat Commun, 15, 2024
|
|
8XR2
 
 | | Crystal structure of AKRtyl-apo1 | | Descriptor: | Aldo/keto reductase | | Authors: | Lin, S, Dai, S, Xiao, Z. | | Deposit date: | 2024-01-06 | | Release date: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | A three-level regulatory mechanism of the aldo-keto reductase subfamily AKR12D
Nat Commun, 15, 2024
|
|
8XQU
 
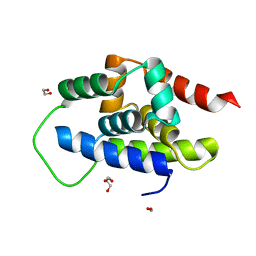 | | The Crystal Structure of ClpC1-NTD from Biortus. | | Descriptor: | 1,2-ETHANEDIOL, ATP-dependent Clp protease ATP-binding subunit ClpC1, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Wang, F, Cheng, W, Lv, Z, Ju, C, Ni, C. | | Deposit date: | 2024-01-05 | | Release date: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | The Crystal Structure of ClpC1-NTD from Biortus.
To Be Published
|
|
8XQK
 
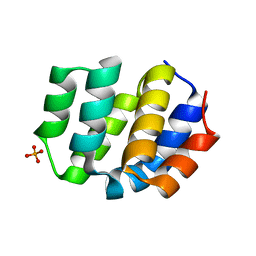 | | The Crystal Structure of Apaf from Biortus. | | Descriptor: | Apoptotic protease-activating factor 1, PHOSPHATE ION | | Authors: | Wang, F, Cheng, W, Lv, Z, Ju, C, Ni, C. | | Deposit date: | 2024-01-05 | | Release date: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | The Crystal Structure of Apaf from Biortus.
To Be Published
|
|
8XQD
 
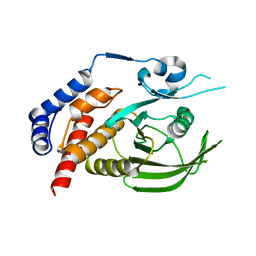 | |
8XQB
 
 | | Mature virion portal vertex of bacteriophage lambda | | Descriptor: | Capsid decoration protein, Head completion protein, Head-tail connector protein FII, ... | | Authors: | Wang, J.W, Gu, Z.W. | | Deposit date: | 2024-01-05 | | Release date: | 2024-04-10 | | Method: | ELECTRON MICROSCOPY (4.07 Å) | | Cite: | Structural morphing in the viral portal vertex of bacteriophage lambda
To Be Published
|
|
