3NX6
 
 | |
1HX5
 
 | |
1G31
 
 | | GP31 CO-CHAPERONIN FROM BACTERIOPHAGE T4 | | Descriptor: | GP31, PHOSPHATE ION, POTASSIUM ION | | Authors: | Hunt, J.F, Van Der Vies, S.M, Henry, L, Deisenhofer, J. | | Deposit date: | 1998-03-27 | | Release date: | 1998-08-26 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural adaptations in the specialized bacteriophage T4 co-chaperonin Gp31 expand the size of the Anfinsen cage.
Cell(Cambridge,Mass.), 90, 1997
|
|
1LEP
 
 | |
1P3H
 
 | | Crystal Structure of the Mycobacterium tuberculosis chaperonin 10 tetradecamer | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 10 kDa chaperonin, CALCIUM ION | | Authors: | Roberts, M.M, Coker, A.R, Fossati, G, Mascagni, P, Coates, A.R.M, Wood, S.P, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2003-04-17 | | Release date: | 2003-07-15 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Mycobacterium tuberculosis chaperonin 10 heptamers self-associate through their biologically active loops
J.BACTERIOL., 185, 2003
|
|
1WNR
 
 | |
7EKQ
 
 | | CrClpP-S2c | | Descriptor: | ATP-dependent Clp protease ATP-binding subunit CLPT4, chloroplastic, ATP-dependent Clp protease proteolytic subunit, ... | | Authors: | Wang, N, Wang, Y.F, Cong, Y, Liu, C.M. | | Deposit date: | 2021-04-06 | | Release date: | 2021-10-20 | | Last modified: | 2022-05-04 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | The cryo-EM structure of the chloroplast ClpP complex.
Nat.Plants, 7, 2021
|
|
4V4O
 
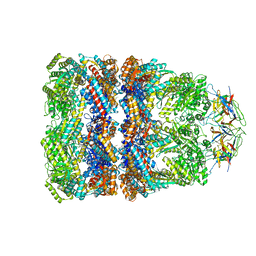 | | Crystal Structure of the Chaperonin Complex Cpn60/Cpn10/(ADP)7 from Thermus Thermophilus | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, DIMETHYL SULFOXIDE, MAGNESIUM ION, ... | | Authors: | Shimamura, T, Koike-Takeshita, A, Yokoyama, K, Masui, R, Murai, N, Yoshida, M, Taguchi, H, Iwata, S. | | Deposit date: | 2004-05-23 | | Release date: | 2014-07-09 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of the native chaperonin complex from Thermus thermophilus revealed unexpected asymmetry at the cis-cavity
STRUCTURE, 12, 2004
|
|
1AON
 
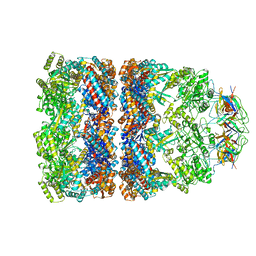 | | CRYSTAL STRUCTURE OF THE ASYMMETRIC CHAPERONIN COMPLEX GROEL/GROES/(ADP)7 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, GROEL, GROEL/GROES COMPLEX, ... | | Authors: | Xu, Z, Horwich, A.L, Sigler, P.B. | | Deposit date: | 1997-07-08 | | Release date: | 1997-10-15 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The crystal structure of the asymmetric GroEL-GroES-(ADP)7 chaperonin complex.
Nature, 388, 1997
|
|
5OPX
 
 | | Crystal structure of the GroEL mutant A109C in complex with GroES and ADP BeF2 | | Descriptor: | 10 kDa chaperonin, 60 kDa chaperonin, ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Yan, X, Shi, Q, Bracher, A, Milicic, G, Singh, A.K, Hartl, F.U, Hayer-Hartl, M. | | Deposit date: | 2017-08-10 | | Release date: | 2018-01-10 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.64 Å) | | Cite: | GroEL Ring Separation and Exchange in the Chaperonin Reaction.
Cell, 172, 2018
|
|
7VWX
 
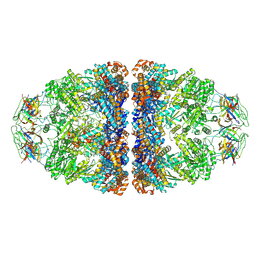 | | CryoEM structure of football-shaped GroEL:ES2 with RuBisCO | | Descriptor: | Chaperonin GroEL, Co-chaperonin GroES, Ribulose bisphosphate carboxylase | | Authors: | Kim, H, Roh, S.H. | | Deposit date: | 2021-11-12 | | Release date: | 2022-01-12 | | Last modified: | 2022-02-16 | | Method: | ELECTRON MICROSCOPY (7.6 Å) | | Cite: | Cryo-EM structures of GroEL:ES 2 with RuBisCO visualize molecular contacts of encapsulated substrates in a double-cage chaperonin.
Iscience, 25, 2022
|
|
8BA9
 
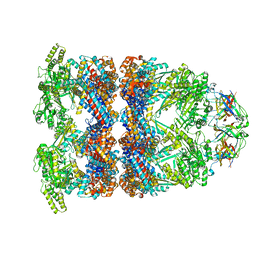 | | CryoEM structure of GroEL-GroES-ADP.AlF3-Rubisco. | | Descriptor: | 60 kDa chaperonin, ADENOSINE-5'-DIPHOSPHATE, ALUMINUM FLUORIDE, ... | | Authors: | Gardner, S, Saibil, H.R. | | Deposit date: | 2022-10-11 | | Release date: | 2023-12-27 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structural basis of substrate progression through the bacterial chaperonin cycle.
Proc Natl Acad Sci U S A, 120, 2023
|
|
3ZQ0
 
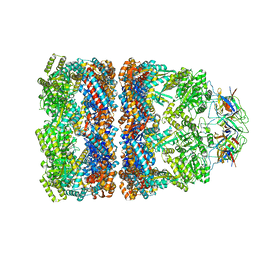 | | Visualizing GroEL-ES in the Act of Encapsulating a Non-Native Substrate Protein | | Descriptor: | 10 KDA CHAPERONIN, 60 KDA CHAPERONIN, ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Chen, D.-H, Madan, D, Weaver, J, Lin, Z, Schroder, G.F, Chiu, W, Rye, H.S. | | Deposit date: | 2013-03-04 | | Release date: | 2013-06-19 | | Last modified: | 2017-08-23 | | Method: | ELECTRON MICROSCOPY (9.2 Å) | | Cite: | Visualizing Groel/Es in the Act of Encapsulating a Folding Protein
Cell(Cambridge,Mass.), 153, 2013
|
|
3ZPZ
 
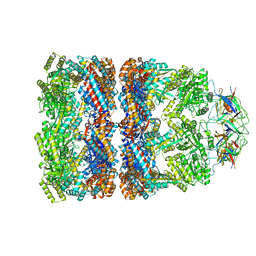 | | Visualizing GroEL-ES in the Act of Encapsulating a Non-Native Substrate Protein | | Descriptor: | 10 KDA CHAPERONIN, 60 KDA CHAPERONIN, ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Chen, D.-H, Madan, D, Weaver, J, Lin, Z, Schroder, G.F, Chiu, W, Rye, H.S. | | Deposit date: | 2013-03-04 | | Release date: | 2013-06-19 | | Last modified: | 2017-08-23 | | Method: | ELECTRON MICROSCOPY (8.9 Å) | | Cite: | Visualizing Groel/Es in the Act of Encapsulating a Folding Protein
Cell(Cambridge,Mass.), 153, 2013
|
|
8G7O
 
 | | ATP- and mtHsp10-bound mtHsp60 V72I focus | | Descriptor: | 10 kDa heat shock protein, mitochondrial, 60 kDa heat shock protein, ... | | Authors: | Braxton, J.R, Shao, H, Tse, E, Gestwicki, J.E, Southworth, D.R. | | Deposit date: | 2023-02-16 | | Release date: | 2023-07-12 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Asymmetric apical domain states of mitochondrial Hsp60 coordinate substrate engagement and chaperonin assembly.
Biorxiv, 2023
|
|
8G7N
 
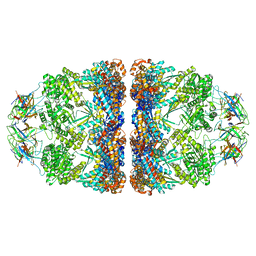 | | ATP- and mtHsp10-bound mtHsp60 V72I | | Descriptor: | 10 kDa heat shock protein, mitochondrial, 60 kDa heat shock protein, ... | | Authors: | Braxton, J.R, Shao, H, Tse, E, Gestwicki, J.E, Southworth, D.R. | | Deposit date: | 2023-02-16 | | Release date: | 2023-07-12 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Asymmetric apical domain states of mitochondrial Hsp60 coordinate substrate engagement and chaperonin assembly.
Biorxiv, 2023
|
|
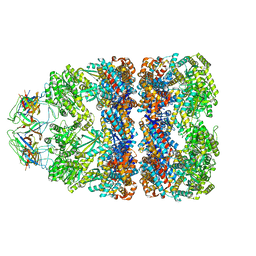
7PBX
 
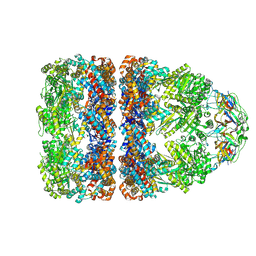 | |
7PBJ
 
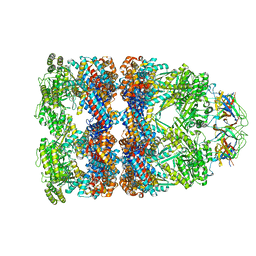 | |
1GRU
 
 | | SOLUTION STRUCTURE OF GROES-ADP7-GROEL-ATP7 COMPLEX BY CRYO-EM | | Descriptor: | GROEL, GROES | | Authors: | Ranson, N.A, Farr, G.W, Roseman, A.M, Gowen, B, Fenton, W.A, Horwich, A.L, Saibil, H.R. | | Deposit date: | 2001-12-16 | | Release date: | 2002-01-28 | | Last modified: | 2019-10-23 | | Method: | ELECTRON MICROSCOPY (12.5 Å) | | Cite: | ATP-Bound States of Groel Captured by Cryo-Electron Microscopy.
Cell(Cambridge,Mass.), 107, 2001
|
|
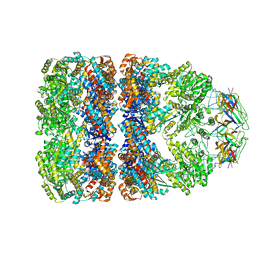
3WVL
 
 | | Crystal structure of the football-shaped GroEL-GroES complex (GroEL: GroES2:ATP14) from Escherichia coli | | Descriptor: | 10 kDa chaperonin, 60 kDa chaperonin, ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Koike-Takeshita, A, Arakawa, T, Taguchi, H, Shimamura, T. | | Deposit date: | 2014-05-23 | | Release date: | 2014-09-17 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.788 Å) | | Cite: | Crystal structure of a symmetric football-shaped GroEL:GroES2-ATP14 complex determined at 3.8 angstrom reveals rearrangement between two GroEL rings.
J.Mol.Biol., 426, 2014
|
|
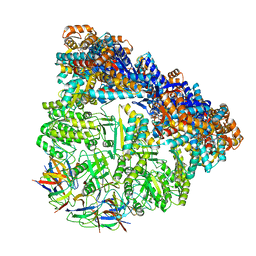
3ZQ1
 
 | | Visualizing GroEL-ES in the Act of Encapsulating a Non-Native Substrate Protein | | Descriptor: | 10 KDA CHAPERONIN, 60 KDA CHAPERONIN, ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Chen, D.-H, Madan, D, Weaver, J, Lin, Z, Schroder, G.F, Chiu, W, Rye, H.S. | | Deposit date: | 2013-03-04 | | Release date: | 2013-06-19 | | Last modified: | 2017-08-23 | | Method: | ELECTRON MICROSCOPY (15.9 Å) | | Cite: | Visualizing Groel/Es in the Act of Encapsulating a Folding Protein
Cell(Cambridge,Mass.), 153, 2013
|
|
1SX4
 
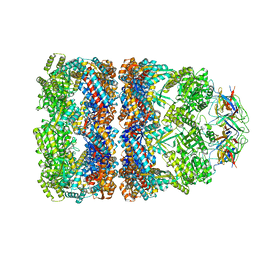 | | GroEL-GroES-ADP7 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, groEL protein, ... | | Authors: | Chaudhry, C, Horwich, A.L, Brunger, A.T, Adams, P.D. | | Deposit date: | 2004-03-30 | | Release date: | 2005-03-01 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Exploring the structural dynamics of the E.coli chaperonin GroEL using translation-libration-screw crystallographic refinement of intermediate states.
J.Mol.Biol., 342, 2004
|
|
8WUW
 
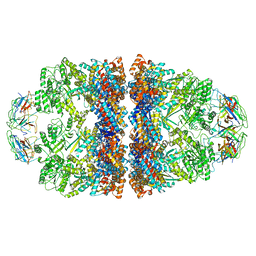 | | Cryo-EM structure of H. thermophilus GroEL-GroES2 asymmetric football complex | | Descriptor: | Chaperonin GroEL, Co-chaperonin GroES, MAGNESIUM ION, ... | | Authors: | Liao, Z, Gopalasingam, C.C, Kameya, M, Gerle, C, Shigematsu, H, Ishii, M, Arakawa, T, Fushinobu, S. | | Deposit date: | 2023-10-21 | | Release date: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Structural insights into thermophilic chaperonin complexes.
Structure, 2024
|
|
8WUX
 
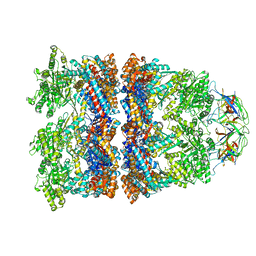 | | Cryo-EM structure of H. thermophilus GroEL-GroES bullet complex | | Descriptor: | Chaperonin GroEL, Co-chaperonin GroES, MAGNESIUM ION, ... | | Authors: | Liao, Z, Gopalasingam, C.C, Kameya, M, Gerle, C, Shigematsu, H, Ishii, M, Arakawa, T, Fushinobu, S. | | Deposit date: | 2023-10-21 | | Release date: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Structural insights into thermophilic chaperonin complexes.
Structure, 2024
|
|
8WUC
 
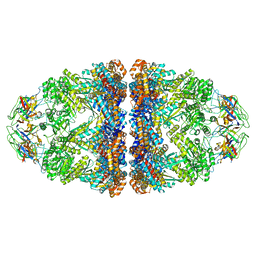 | | Cryo-EM structure of H. thermoluteolus GroEL-GroES2 football complex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Chaperonin GroEL, Co-chaperonin GroES, ... | | Authors: | Liao, Z, Gopalasingam, C.C, Kameya, M, Gerle, C, Shigematsu, H, Ishii, M, Arakawa, T, Fushinobu, S. | | Deposit date: | 2023-10-20 | | Release date: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Structural insights into thermophilic chaperonin complexes.
Structure, 2024
|
|
