6XRO
 
 | |
4H1D
 
 | | Cocrystal structure of GlpG and DFP | | Descriptor: | DIISOPROPYL PHOSPHONATE, Rhomboid protease GlpG | | Authors: | Xue, Y, Ha, Y. | | Deposit date: | 2012-09-10 | | Release date: | 2013-05-01 | | Last modified: | 2013-06-26 | | Method: | X-RAY DIFFRACTION (2.8975 Å) | | Cite: | Large lateral movement of transmembrane helix s5 is not required for substrate access to the active site of rhomboid intramembrane protease.
J.Biol.Chem., 288, 2013
|
|
3ODJ
 
 | | Crystal structure of H. influenzae rhomboid GlpG with disordered loop 4, helix 5 and loop 5 | | Descriptor: | Rhomboid protease glpG | | Authors: | Brooks, C.L, Lazareno-Saez, C, Lamoureux, J.S, Mak, M.W, Lemieux, M.J. | | Deposit date: | 2010-08-11 | | Release date: | 2011-02-23 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.84 Å) | | Cite: | Insights into Substrate Gating in H. influenzae Rhomboid.
J.Mol.Biol., 407, 2011
|
|
8AB5
 
 | | Structure of E. coli GlpG in complex with peptide derived inhibitor Ac-VRHA-conh-[4-(4-butyl)-phenoxy-1-phenyl-2-butyl] | | Descriptor: | Ac-VRHA-conh-[4-(4-butyl)-phenoxy-1-phenyl-2-butyl], Rhomboid protease GlpG | | Authors: | Skerlova, J, Polovinkin, V, Bach, K, Borshchevskiy, V, Strisovsky, K. | | Deposit date: | 2022-07-04 | | Release date: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of E. coli GlpG in complex with peptide derived inhibitor Ac-VRHA-conh-[4-(4-butyl)-phenoxy-1-phenyl-2-butyl]
To Be Published
|
|
3B44
 
 | | Crystal structure of GlpG W136A mutant | | Descriptor: | glpG, nonyl beta-D-glucopyranoside | | Authors: | Wang, Y, Maegawa, S, Akiyama, Y, Ha, Y. | | Deposit date: | 2007-10-23 | | Release date: | 2008-01-22 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The role of L1 loop in the mechanism of rhomboid intramembrane protease GlpG.
J.Mol.Biol., 374, 2007
|
|
3B45
 
 | | Crystal structure of GlpG at 1.9A resolution | | Descriptor: | glpG, nonyl beta-D-glucopyranoside | | Authors: | Wang, Y, Maegawa, S, Akiyama, Y, Ha, Y. | | Deposit date: | 2007-10-23 | | Release date: | 2008-01-22 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The role of L1 loop in the mechanism of rhomboid intramembrane protease GlpG.
J.Mol.Biol., 374, 2007
|
|
4NJN
 
 | | Crystal Structure of E.coli GlpG at pH 4.5 | | Descriptor: | Rhomboid protease GlpG | | Authors: | Dickey, S.W, Baker, R.P, Cho, S, Urban, S. | | Deposit date: | 2013-11-11 | | Release date: | 2013-12-25 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Proteolysis inside the Membrane Is a Rate-Governed Reaction Not Driven by Substrate Affinity.
Cell(Cambridge,Mass.), 155, 2013
|
|
4NJP
 
 | | Proteolysis inside the membrane is a rate-governed reaction not Driven by substrate affinity | | Descriptor: | Rhomboid protease GlpG | | Authors: | Dickey, S.W, Baker, R.P, Cho, S, Urban, S. | | Deposit date: | 2013-11-11 | | Release date: | 2013-12-25 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Proteolysis inside the Membrane Is a Rate-Governed Reaction Not Driven by Substrate Affinity.
Cell(Cambridge,Mass.), 155, 2013
|
|
2IRV
 
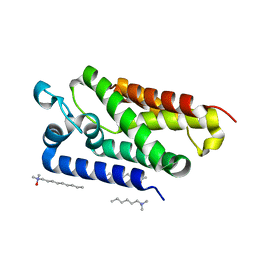 | | Crystal structure of GlpG, a rhomboid intramembrane serine protease | | Descriptor: | (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (11E)-OCTADEC-11-ENOATE, DODECYL-BETA-D-MALTOSIDE, LAURYL DIMETHYLAMINE-N-OXIDE, ... | | Authors: | Bibi, E, Fass, D, Ben-Shem, A. | | Deposit date: | 2006-10-16 | | Release date: | 2006-10-31 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis for intramembrane proteolysis by rhomboid serine proteases.
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
2IC8
 
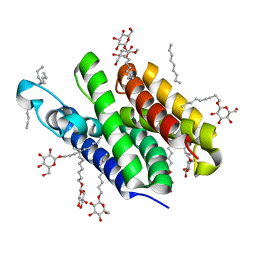 | | Crystal structure of GlpG | | Descriptor: | Protein glpG, nonyl beta-D-glucopyranoside | | Authors: | Ha, Y. | | Deposit date: | 2006-09-12 | | Release date: | 2006-10-17 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of a rhomboid family intramembrane protease.
Nature, 444, 2006
|
|
6VJ8
 
 | |
6VJ9
 
 | |
2NRF
 
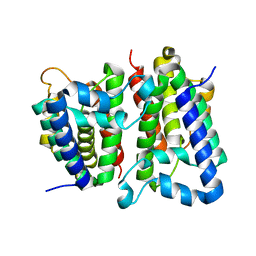 | | Crystal Structure of GlpG, a Rhomboid family intramembrane protease | | Descriptor: | Protein GlpG | | Authors: | Wu, Z, Yan, N, Feng, L, Yan, H, Gu, L, Shi, Y. | | Deposit date: | 2006-11-02 | | Release date: | 2006-11-14 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural analysis of a rhomboid family intramembrane protease reveals a gating mechanism for substrate entry.
Nat.Struct.Mol.Biol., 13, 2006
|
|
2NR9
 
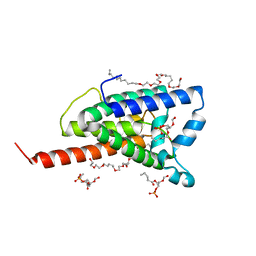 | | Crystal structure of GlpG, Rhomboid Peptidase from Haemophilus influenzae | | Descriptor: | (R)-2-(FORMYLOXY)-3-(PHOSPHONOOXY)PROPYL PENTANOATE, 3,6,12,15,18,21,24-HEPTAOXAHEXATRIACONTAN-1-OL, Protein glpG homolog | | Authors: | Lemieux, M.J, Fischer, S.J, Cherney, M.M, Bateman, K.S, James, M.N.G. | | Deposit date: | 2006-11-01 | | Release date: | 2006-11-14 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The crystal structure of the rhomboid peptidase from Haemophilus influenzae provides insight into intramembrane proteolysis.
Proc.Natl.Acad.Sci.USA, 104, 2007
|
|
2O7L
 
 | | The open-cap conformation of GlpG | | Descriptor: | Protein glpG, nonyl beta-D-glucopyranoside | | Authors: | Ha, Y. | | Deposit date: | 2006-12-11 | | Release date: | 2006-12-26 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Open-cap conformation of intramembrane protease GlpG.
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
6XRP
 
 | |
5MT7
 
 | |
2XOW
 
 | | Structure of GlpG in complex with a mechanism-based isocoumarin inhibitor | | Descriptor: | 5-AMINO-2-(2-METHOXY-2-OXOETHYL)BENZOIC ACID, RHOMBOID PROTEASE GLPG, nonyl beta-D-glucopyranoside | | Authors: | Vinothkumar, K.R, Strisovsky, K, Andreeva, A, Christova, Y, Verhelst, S, Freeman, M. | | Deposit date: | 2010-08-24 | | Release date: | 2010-10-13 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | The Structural Basis for Catalysis and Substrate Specificity of a Rhomboid Protease
Embo J., 29, 2010
|
|
5MT6
 
 | |
2XOV
 
 | | Crystal Structure of E.coli rhomboid protease GlpG, native enzyme | | Descriptor: | GLYCEROL, RHOMBOID PROTEASE GLPG, nonyl beta-D-glucopyranoside | | Authors: | Vinothkumar, K.R, Strisovsky, K, Andreeva, A, Christova, Y, Verhelst, S, Freeman, M. | | Deposit date: | 2010-08-24 | | Release date: | 2010-10-13 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | The structural basis for catalysis and substrate specificity of a rhomboid protease.
EMBO J., 29, 2010
|
|
2XTU
 
 | |
5MT8
 
 | |
5MTF
 
 | | A modular route to novel potent and selective inhibitors of rhomboid intramembrane proteases | | Descriptor: | CHLORIDE ION, Rhomboid protease GlpG, inhibitor, ... | | Authors: | Ticha, A, Stanchev, S, Vinothkumar, K.R, Mikles, D.C, Pachl, P, Svehlova, K, Nguyen, M.T.N, Verhelst, S.H.L, Johnson, D, Bachovchin, D, Lepsik, M, Majer, P, Strisovsky, K. | | Deposit date: | 2017-01-09 | | Release date: | 2017-11-15 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | General and Modular Strategy for Designing Potent, Selective, and Pharmacologically Compliant Inhibitors of Rhomboid Proteases.
Cell Chem Biol, 24, 2017
|
|
2XTV
 
 | |
5F5B
 
 | |
