5O6F
 
 | | NMR structure of cold shock protein A from Corynebacterium pseudotuberculosis | | Descriptor: | Cold-shock protein | | Authors: | Caruso, I.P, Panwalkar, V, Coronado, M.A, Dingley, A.J, Cornelio, M.L, Willbold, D, Arni, R.K, Eberle, R.J. | | Deposit date: | 2017-06-06 | | Release date: | 2017-07-19 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Structure and interaction of Corynebacterium pseudotuberculosis cold shock protein A with Y-box single-stranded DNA fragment.
FEBS J., 285, 2018
|
|
1C9O
 
 | | CRYSTAL STRUCTURE ANALYSIS OF THE BACILLUS CALDOLYTICUS COLD SHOCK PROTEIN BC-CSP | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, COLD-SHOCK PROTEIN, SODIUM ION | | Authors: | Mueller, U, Perl, D, Schmid, F.X, Heinemann, U. | | Deposit date: | 1999-08-03 | | Release date: | 2000-04-02 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.17 Å) | | Cite: | Thermal stability and atomic-resolution crystal structure of the Bacillus caldolyticus cold shock protein.
J.Mol.Biol., 297, 2000
|
|
3I2Z
 
 | | Structure of cold shock protein E from Salmonella typhimurium | | Descriptor: | RNA chaperone, negative regulator of cspA transcription | | Authors: | Morgan, H.P, McNae, I, Wear, M.A, Gallagher, M, Walkinshaw, M.D. | | Deposit date: | 2009-06-30 | | Release date: | 2009-12-22 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Crystallization and X-ray structure of cold-shock protein E from Salmonella typhimurium
Acta Crystallogr.,Sect.F, 65, 2009
|
|
2BH8
 
 | | Combinatorial Protein 1b11 | | Descriptor: | 1B11 | | Authors: | De Bono, S, Riechmann, L, Girard, E, Williams, R.L, Winter, G. | | Deposit date: | 2005-01-07 | | Release date: | 2005-02-07 | | Last modified: | 2018-01-24 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A Segment of Cold Shock Protein Directs the Folding of a Combinatorial Protein
Proc.Natl.Acad.Sci.USA, 102, 2005
|
|
5JX4
 
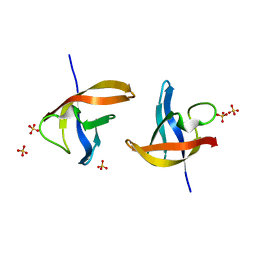 | | Crystal structure of E36-G37del mutant of the Bacillus caldolyticus cold shock protein. | | Descriptor: | Cold shock protein CspB, SULFATE ION | | Authors: | Carvajal, A, Castro-Fernandez, V, Cabrejos, D, Fuentealba, M, Pereira, H.M, Vallejos, G, Cabrera, R, Garratt, R.C, Komives, E.A, Ramirez-Sarmiento, C.A, Babul, J. | | Deposit date: | 2016-05-12 | | Release date: | 2017-05-10 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Unusual dimerization of a BcCsp mutant leads to reduced conformational dynamics.
FEBS J., 284, 2017
|
|
1NMF
 
 | |
1NMG
 
 | |
6SZZ
 
 | |
6T00
 
 | |
7ZHH
 
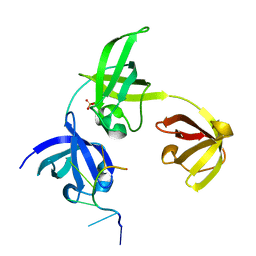 | | Complex structure of drosophila Unr CSD789 and a poly(A) RNA sequence | | Descriptor: | RNA (5'-R(P*AP*AP*AP*AP*AP*A)-3'), SULFATE ION, Upstream of N-ras, ... | | Authors: | Hollmann, N.M, Jagtap, P.K.A, Hennig, J. | | Deposit date: | 2022-04-06 | | Release date: | 2022-12-07 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Upstream of N-Ras C-terminal cold shock domains mediate poly(A) specificity in a novel RNA recognition mode and bind poly(A) binding protein.
Nucleic Acids Res., 51, 2023
|
|
1WFQ
 
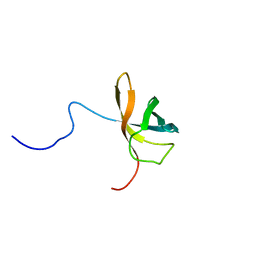 | | Solution structure of the first cold-shock domain of the human KIAA0885 protein (UNR protein) | | Descriptor: | UNR protein | | Authors: | Goroncy, A.K, Kigawa, T, Koshiba, S, Tomizawa, T, Kobayashi, N, Tochio, N, Inoue, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-05-26 | | Release date: | 2004-11-26 | | Last modified: | 2011-07-13 | | Method: | SOLUTION NMR | | Cite: | The NMR solution structures of the five constituent cold-shock domains (CSD) of the human UNR (upstream of N-ras) protein.
J.Struct.Funct.Genom., 11, 2010
|
|
1X65
 
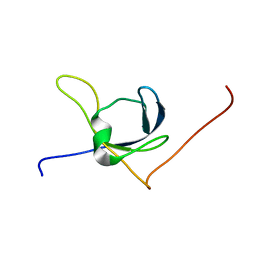 | | Solution structure of the third cold-shock domain of the human KIAA0885 protein (UNR PROTEIN) | | Descriptor: | UNR protein | | Authors: | Goroncy, A.K, Kigawa, T, Koshiba, S, Kobayashi, N, Tochio, N, Inoue, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-05-17 | | Release date: | 2005-11-17 | | Last modified: | 2011-07-13 | | Method: | SOLUTION NMR | | Cite: | The NMR solution structures of the five constituent cold-shock domains (CSD) of the human UNR (upstream of N-ras) protein.
J.Struct.Funct.Genom., 11, 2010
|
|
3A0J
 
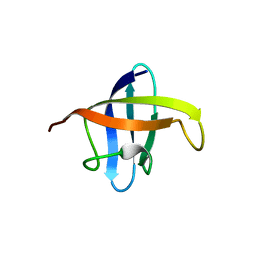 | | Crystal structure of cold shock protein 1 from Thermus thermophilus HB8 | | Descriptor: | Cold shock protein | | Authors: | Miyazaki, T, Nakagawa, N, Kuramitsu, S, Masui, R, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2009-03-19 | | Release date: | 2010-03-23 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | The Biological Action of Cold Shock Protein 1 from Thermus thermophilus HB8
To be Published
|
|
3MEF
 
 | |
3AQQ
 
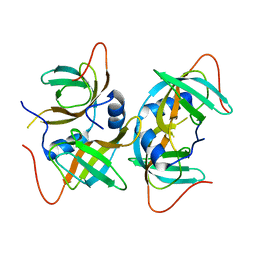 | | Crystal structure of human CRHSP-24 | | Descriptor: | Calcium-regulated heat stable protein 1 | | Authors: | Hou, H, Wang, F, Zhang, W, Wang, D, Li, X, Bartlam, M, Yao, X, Rao, Z. | | Deposit date: | 2010-11-17 | | Release date: | 2010-12-22 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.798 Å) | | Cite: | CRHSP-24 is a novel cargo adaptor trafficking between stress granules and processing bodies
To be Published
|
|
3CAM
 
 | |
2YTY
 
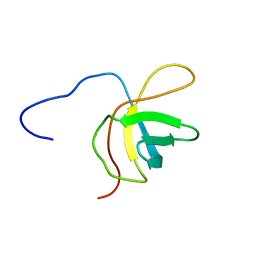 | | Solution structure of the fourth cold-shock domain of the human KIAA0885 protein (UNR protein) | | Descriptor: | Cold shock domain-containing protein E1 | | Authors: | Goroncy, A.K, Tomizawa, T, Koshiba, S, Inoue, M, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-04-05 | | Release date: | 2008-04-08 | | Last modified: | 2011-07-13 | | Method: | SOLUTION NMR | | Cite: | The NMR solution structures of the five constituent cold-shock domains (CSD) of the human UNR (upstream of N-ras) protein.
J.Struct.Funct.Genom., 11, 2010
|
|
2YTX
 
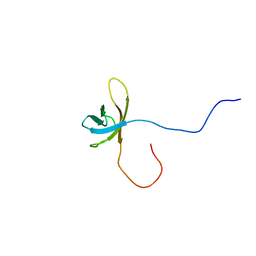 | | Solution structure of the second cold-shock domain of the human KIAA0885 protein (UNR protein) | | Descriptor: | Cold shock domain-containing protein E1 | | Authors: | Goroncy, A.K, Tomizawa, T, Koshiba, S, Inoue, M, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-04-05 | | Release date: | 2008-04-08 | | Last modified: | 2011-07-13 | | Method: | SOLUTION NMR | | Cite: | The NMR solution structures of the five constituent cold-shock domains (CSD) of the human UNR (upstream of N-ras) protein.
J.Struct.Funct.Genom., 11, 2010
|
|
2YTV
 
 | | Solution structure of the fifth cold-shock domain of the human KIAA0885 protein (unr protein) | | Descriptor: | Cold shock domain-containing protein E1 | | Authors: | Goroncy, A.K, Tochio, N, Tomizawa, T, Koshiba, S, Watanabe, S, Harada, T, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-04-05 | | Release date: | 2008-04-08 | | Last modified: | 2011-07-13 | | Method: | SOLUTION NMR | | Cite: | The NMR solution structures of the five constituent cold-shock domains (CSD) of the human UNR (upstream of N-ras) protein.
J.Struct.Funct.Genom., 11, 2010
|
|
3PF4
 
 | | Crystal structure of Bs-CspB in complex with r(GUCUUUA) | | Descriptor: | Cold shock protein cspB, MAGNESIUM ION, SODIUM ION, ... | | Authors: | Sachs, R, Max, K.E.A, Heinemann, U. | | Deposit date: | 2010-10-27 | | Release date: | 2011-09-21 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | RNA single strands bind to a conserved surface of the major cold shock protein in crystals and solution.
Rna, 18, 2012
|
|
3PF5
 
 | |
1CSQ
 
 | |
1CSP
 
 | |
2LXJ
 
 | | Backbone 1H, 13C, and 15N Chemical Shift Assignments for cold shock protein, LmCsp with dT7 | | Descriptor: | Cold shock-like protein CspLA | | Authors: | Lee, J, Jeong, K, Kim, Y. | | Deposit date: | 2012-08-27 | | Release date: | 2013-08-07 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Structural and Dynamic Features of Cold-Shock Proteins of Listeriamonocytogenes, a Psychrophilic Bacterium
Biochemistry, 52, 2013
|
|
4A4I
 
 | | Crystal structure of the human Lin28b cold shock domain | | Descriptor: | GLYCEROL, PROTEIN LIN-28 HOMOLOG B, SULFATE ION | | Authors: | Mayr, F, Schuetz, A, Doege, N, Heinemann, U. | | Deposit date: | 2011-10-14 | | Release date: | 2012-08-08 | | Last modified: | 2012-09-05 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | The Lin28 Cold-Shock Domain Remodels Pre-Let-7 Microrna.
Nucleic Acids Res., 40, 2012
|
|
