4WCZ
 
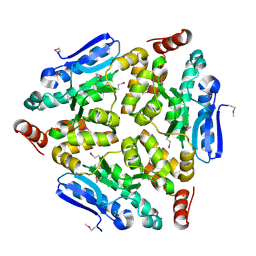 | | Crystal structure of a putative enoyl-CoA hydratase/isomerase from Novosphingobium aromaticivorans | | Descriptor: | Enoyl-CoA hydratase/isomerase | | Authors: | Tkaczuk, K.L, Cooper, D.R, Chapman, H.C, Niedzialkowska, E, Cymborowski, M.T, Hillerich, B.S, Stead, M, Ahmed, M, Hammonds, J, Bonanno, J, Seidel, R, Almo, S.C, Minor, W, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2014-09-05 | | Release date: | 2014-12-03 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Crystal structure of a putative enoyl-CoA hydratase/isomerase from Novosphingobium aromaticivorans
to be published
|
|
6WYI
 
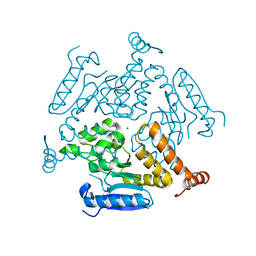 | | Crystal structure of EchA19, enoyl-CoA hydratase from Mycobacterium tuberculosis | | Descriptor: | EchA19, enoyl-CoA hydratase, MAGNESIUM ION | | Authors: | Bonds, A.C, Garcia-Diaz, M, Sampson, N.S. | | Deposit date: | 2020-05-13 | | Release date: | 2020-07-29 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.915 Å) | | Cite: | Post-translational Succinylation ofMycobacterium tuberculosisEnoyl-CoA Hydratase EchA19 Slows Catalytic Hydration of Cholesterol Catabolite 3-Oxo-chol-4,22-diene-24-oyl-CoA.
Acs Infect Dis., 6, 2020
|
|
8SK6
 
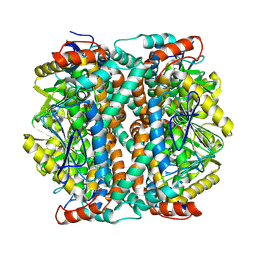 | | human liver mitochondrial Delta(3,5)-Delta(2,4)-dienoyl-CoA isomerase | | Descriptor: | Delta(3,5)-Delta(2,4)-dienoyl-CoA isomerase, mitochondrial | | Authors: | Zhang, Z, Tringides, M. | | Deposit date: | 2023-04-18 | | Release date: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (2.96 Å) | | Cite: | High-Resolution Structural Proteomics of Mitochondria Using the 'Build and Retrieve' Methodology.
Mol.Cell Proteomics, 22, 2023
|
|
8UJA
 
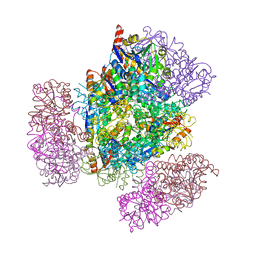 | |
5O34
 
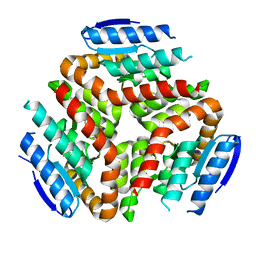 | | ThnE from S.clavuligerus | | Descriptor: | Enoyl-CoA hydratase carB homologue | | Authors: | Valegard, K, Andersson, I. | | Deposit date: | 2017-05-23 | | Release date: | 2018-06-13 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Structure of ThnE from the thienamycin biosynthesis.
To Be Published
|
|
2ZQQ
 
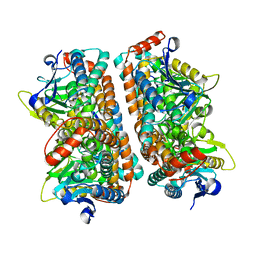 | | Crystal structure of human AUH (3-methylglutaconyl-coa hydratase) mixed with (AUUU)24A RNA | | Descriptor: | Methylglutaconyl-CoA hydratase | | Authors: | Kurimoto, K, Kuwasako, K, Muto, Y, Nureki, O, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2008-08-16 | | Release date: | 2009-06-30 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | AU-rich RNA-binding induces changes in the quaternary structure of AUH
Proteins, 75, 2009
|
|
2ZQR
 
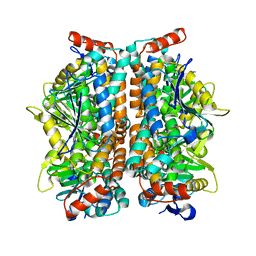 | | Crystal structure of AUH without RNA | | Descriptor: | Methylglutaconyl-CoA hydratase | | Authors: | Kurimoto, K, Kuwasako, K, Muto, Y, Nureki, O, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2008-08-16 | | Release date: | 2009-06-30 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | AU-rich RNA-binding induces changes in the quaternary structure of AUH
Proteins, 75, 2009
|
|
7BOR
 
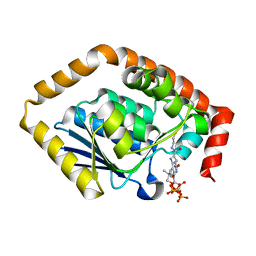 | | Structure of Pseudomonas aeruginosa CoA-bound OdaA | | Descriptor: | COENZYME A, Probable enoyl-CoA hydratase/isomerase | | Authors: | Zhao, N, Zhao, C, Liu, L, Li, T, Li, C, He, L, Zhu, Y, Song, Y, Bao, R. | | Deposit date: | 2020-03-19 | | Release date: | 2020-05-13 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.901 Å) | | Cite: | Structural and molecular dynamic studies of Pseudomonas aeruginosa OdaA reveal the regulation role of a C-terminal hinge element.
Biochim Biophys Acta Gen Subj, 1865, 2020
|
|
7CRD
 
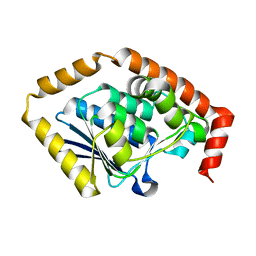 | | Structure of Pseudomonas aeruginosa OdaA | | Descriptor: | Probable enoyl-CoA hydratase/isomerase | | Authors: | Zhao, N, Zhao, C, Liu, L, Li, T, Li, C, He, L, Zhu, Y, Song, Y, Bao, R. | | Deposit date: | 2020-08-13 | | Release date: | 2020-09-16 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.901 Å) | | Cite: | Structural and molecular dynamic studies of Pseudomonas aeruginosa OdaA reveal the regulation role of a C-terminal hinge element.
Biochim Biophys Acta Gen Subj, 1865, 2020
|
|
5WYB
 
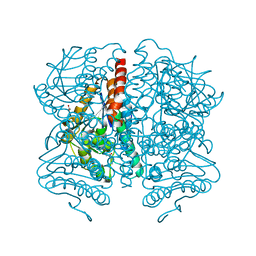 | | Structure of Pseudomonas aeruginosa DspI | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, ACETATE ION, Probable enoyl-CoA hydratase/isomerase | | Authors: | Liu, L, Peng, C, Li, T, Li, C, He, L, Song, Y, Zhu, Y, Shen, Y, Bao, R. | | Deposit date: | 2017-01-12 | | Release date: | 2018-01-17 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural and functional studies on Pseudomonas aeruginosa DspI: implications for its role in DSF biosynthesis.
Sci Rep, 8, 2018
|
|
5WYD
 
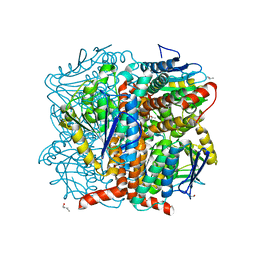 | | Structural of Pseudomonas aeruginosa DspI | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, ISOPROPYL ALCOHOL, ... | | Authors: | Liu, L, Peng, C, Li, T, Li, C, He, L, Song, Y, Zhu, Y, Shen, Y, Bao, R. | | Deposit date: | 2017-01-12 | | Release date: | 2018-01-31 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.101 Å) | | Cite: | Structural and functional studies on Pseudomonas aeruginosa DspI: implications for its role in DSF biosynthesis.
Sci Rep, 8, 2018
|
|
7EUM
 
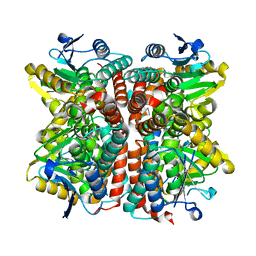 | |
6DGN
 
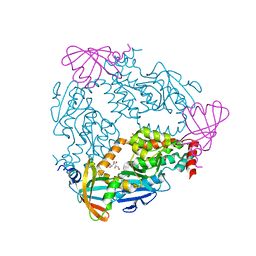 | |
5JBX
 
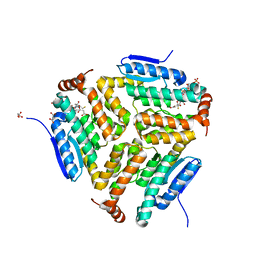 | | Crystal structure of LiuC in complex with coenzyme A and malonic acid | | Descriptor: | 3-hydroxybutyryl-CoA dehydratase, COENZYME A, MALONATE ION | | Authors: | Bock, T, Reichelt, J, Mueller, R, Blankenfeldt, W. | | Deposit date: | 2016-04-14 | | Release date: | 2016-06-22 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | The Structure of LiuC, a 3-Hydroxy-3-Methylglutaconyl CoA Dehydratase Involved in Isovaleryl-CoA Biosynthesis in Myxococcus xanthus, Reveals Insights into Specificity and Catalysis.
Chembiochem, 17, 2016
|
|
5JBW
 
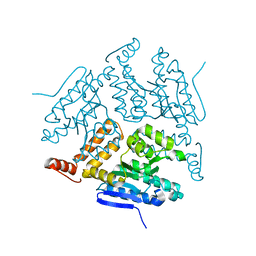 | | Crystal structure of LiuC | | Descriptor: | 3-hydroxybutyryl-CoA dehydratase | | Authors: | Bock, T, Reichelt, J, Mueller, R, Blankenfeldt, W. | | Deposit date: | 2016-04-14 | | Release date: | 2016-06-22 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | The Structure of LiuC, a 3-Hydroxy-3-Methylglutaconyl CoA Dehydratase Involved in Isovaleryl-CoA Biosynthesis in Myxococcus xanthus, Reveals Insights into Specificity and Catalysis.
Chembiochem, 17, 2016
|
|
5KAJ
 
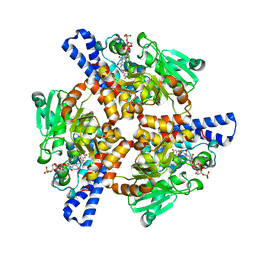 | | Crystal structure of a dioxygenase in the Crotonase superfamily in P21, A319C mutant | | Descriptor: | (3,5-dihydroxyphenyl)acetyl-CoA 1,2-dioxygenase, [[(2~{R},3~{S},4~{R},5~{R})-5-(6-aminopurin-9-yl)-4-oxidanyl-3-phosphonooxy-oxolan-2-yl]methoxy-oxidanyl-phosphoryl] [(3~{R})-4-[[3-[2-[(~{E})-2-[3,5-bis(oxidanyl)phenyl]-1-oxidanyl-ethenyl]sulfanylethylamino]-3-oxidanylidene-propyl]amino]-2,2-dimethyl-3-oxidanyl-4-oxidanylidene-butyl] hydrogen phosphate | | Authors: | Li, K, Fielding, E.N, Condurso, H.L, Bruner, S.D. | | Deposit date: | 2016-06-01 | | Release date: | 2017-06-21 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.681 Å) | | Cite: | Probing the structural basis of oxygen binding in a cofactor-independent dioxygenase.
Acta Crystallogr D Struct Biol, 73, 2017
|
|
7M3W
 
 | |
7MCM
 
 | |
6N92
 
 | | Methylmalonyl-CoA decarboxylase in complex with 2-nitronate-propionyl-CoA | | Descriptor: | (2E)-2-(hydroxyimino)propanoic acid, IMIDAZOLE, Methylmalonyl-CoA decarboxylase, ... | | Authors: | Stunkard, L.M, Dixon, A.D, Huth, T.J, Lohman, J.R. | | Deposit date: | 2018-11-30 | | Release date: | 2019-04-10 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Sulfonate/Nitro Bearing Methylmalonyl-Thioester Isosteres Applied to Methylmalonyl-CoA Decarboxylase Structure-Function Studies.
J. Am. Chem. Soc., 141, 2019
|
|
6N97
 
 | | Methylmalonyl-CoA decarboxylase in complex with 2-sulfonate-propionyl-amino(dethia)-CoA | | Descriptor: | (2R)-sulfonatepropionyl-amino(dethia)-CoA, (2S)-sulfonatepropionyl-amino(dethia)-CoA, IMIDAZOLE, ... | | Authors: | Stunkard, L.M, Dixon, A.D, Huth, T.J, Lohman, J.R. | | Deposit date: | 2018-11-30 | | Release date: | 2019-04-10 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Sulfonate/Nitro Bearing Methylmalonyl-Thioester Isosteres Applied to Methylmalonyl-CoA Decarboxylase Structure-Function Studies.
J. Am. Chem. Soc., 141, 2019
|
|
5KAH
 
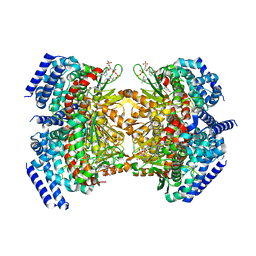 | | Crystal structure of a dioxygenase in the Crotonase superfamily in P21, V425T mutant | | Descriptor: | (3,5-dihydroxyphenyl)acetyl-CoA 1,2-dioxygenase, [(2R,3S,4R,5R)-5-(6-AMINO-9H-PURIN-9-YL)-4-HYDROXY-3-(PHOSPHONOOXY)TETRAHYDROFURAN-2-YL]METHYL (3R)-4-({3-[(2-{[(3,5-DIHYDROXYPHENYL)ACETYL]AMINO}ETHYL)AMINO]-3-OXOPROPYL}AMINO)-3-HYDROXY-2,2-DIMETHYL-4-OXOBUTYL DIHYDROGEN DIPHOSPHATE | | Authors: | Li, K, Fielding, E.N, Condurso, H.L, Bruner, S.D. | | Deposit date: | 2016-06-01 | | Release date: | 2017-06-21 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.779 Å) | | Cite: | Probing the structural basis of oxygen binding in a cofactor-independent dioxygenase.
Acta Crystallogr D Struct Biol, 73, 2017
|
|
5KAG
 
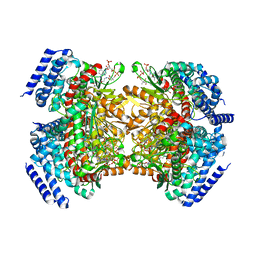 | | Crystal structure of a dioxygenase in the Crotonase superfamily in P21 | | Descriptor: | (3,5-dihydroxyphenyl)acetyl-CoA 1,2-dioxygenase, OXYGEN MOLECULE, [(2R,3S,4R,5R)-5-(6-AMINO-9H-PURIN-9-YL)-4-HYDROXY-3-(PHOSPHONOOXY)TETRAHYDROFURAN-2-YL]METHYL (3R)-4-({3-[(2-{[(3,5-DIHYDROXYPHENYL)ACETYL]AMINO}ETHYL)AMINO]-3-OXOPROPYL}AMINO)-3-HYDROXY-2,2-DIMETHYL-4-OXOBUTYL DIHYDROGEN DIPHOSPHATE | | Authors: | Li, K, Fielding, E.N, Condurso, H.L, Bruner, S.D. | | Deposit date: | 2016-06-01 | | Release date: | 2017-06-21 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.456 Å) | | Cite: | Probing the structural basis of oxygen binding in a cofactor-independent dioxygenase.
Acta Crystallogr D Struct Biol, 73, 2017
|
|
1XX4
 
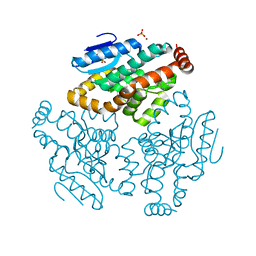 | | Crystal Structure of Rat Mitochondrial 3,2-Enoyl-CoA | | Descriptor: | 3,2-trans-enoyl-CoA isomerase, mitochondrial, BENZAMIDINE, ... | | Authors: | Hubbard, P.A, Yu, W, Schulz, H, Kim, J.-J. | | Deposit date: | 2004-11-03 | | Release date: | 2004-11-23 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Domain swapping in the low-similarity isomerase/hydratase superfamily: the crystal structure of rat mitochondrial Delta3, Delta2-enoyl-CoA isomerase.
Protein Sci., 14, 2005
|
|
5KJP
 
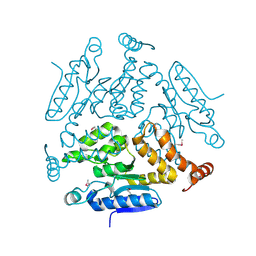 | |
6OJM
 
 | |
