7R0D
 
 | |
8SXS
 
 | |
1F3Y
 
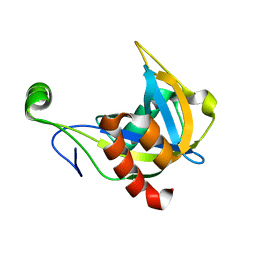 | | SOLUTION STRUCTURE OF THE NUDIX ENZYME DIADENOSINE TETRAPHOSPHATE HYDROLASE FROM LUPINUS ANGUSTIFOLIUS L. | | Descriptor: | DIADENOSINE 5',5'''-P1,P4-TETRAPHOSPHATE HYDROLASE | | Authors: | Swarbrick, J.D, Bashtannyk, T, Maksel, D, Zhang, X.R, Blackburn, G.M, Gayler, K.R, Gooley, P.R. | | Deposit date: | 2000-06-06 | | Release date: | 2001-06-06 | | Last modified: | 2021-11-10 | | Method: | SOLUTION NMR | | Cite: | The three-dimensional structure of the Nudix enzyme diadenosine tetraphosphate hydrolase from Lupinus angustifolius L.
J.Mol.Biol., 302, 2000
|
|
6QVO
 
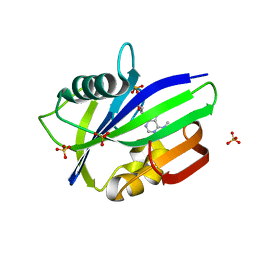 | | Crystal structure of human MTH1 in complex with N6-methyl-dAMP | | Descriptor: | 7,8-dihydro-8-oxoguanine triphosphatase, GLYCEROL, N6-METHYL-DEOXY-ADENOSINE-5'-MONOPHOSPHATE, ... | | Authors: | Scaletti, E, Vallin, K.S, Brautigam, L, Sarno, A, Warpman Berglund, U, Helleday, T, Stenmark, P, Jemth, A.S. | | Deposit date: | 2019-03-04 | | Release date: | 2020-03-18 | | Last modified: | 2020-09-30 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | MutT homologue 1 (MTH1) removes N6-methyl-dATP from the dNTP pool.
J.Biol.Chem., 295, 2020
|
|
8DP8
 
 | |
8DPA
 
 | | Crystal structure of the homodimeric AvrM14-B Nudix hydrolase effector from Melampsora lini | | Descriptor: | AvrM14-B, SULFATE ION | | Authors: | McCombe, C.L, Outram, M.A, Ericsson, D.J, Williams, S.J. | | Deposit date: | 2022-07-15 | | Release date: | 2023-01-25 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | A rust-fungus Nudix hydrolase effector decaps mRNA in vitro and interferes with plant immune pathways.
New Phytol., 239, 2023
|
|
8DP9
 
 | | Crystal structure of the monomeric AvrM14-B Nudix hydrolase effector from Melampsora lini | | Descriptor: | AvrM14-B, SULFATE ION | | Authors: | McCombe, C.L, Outram, M.A, Ericsson, D.J, Williams, S.J. | | Deposit date: | 2022-07-15 | | Release date: | 2023-01-25 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | A rust-fungus Nudix hydrolase effector decaps mRNA in vitro and interferes with plant immune pathways.
New Phytol., 239, 2023
|
|
6T5J
 
 | | Structure of NUDT15 in complex with inhibitor TH1760 | | Descriptor: | 6-[4-(1~{H}-indol-5-ylcarbonyl)piperazin-1-yl]sulfonyl-3~{H}-1,3-benzoxazol-2-one, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Carter, M, Rehling, D, Desroses, M, Zhang, S.M, Hagenkort, A, Valerie, N.C.K, Helleday, T, Stenmark, P. | | Deposit date: | 2019-10-16 | | Release date: | 2020-07-29 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Development of a chemical probe against NUDT15.
Nat.Chem.Biol., 16, 2020
|
|
7N13
 
 | | Crystal structure of MTH1 in complex with compound 32 | | Descriptor: | 4-anilino-6-[4-(butylcarbamoyl)-3-fluorophenyl]-N-cyclopropyl-7-fluoroquinoline-3-carboxamide, 7,8-dihydro-8-oxoguanine triphosphatase, SULFATE ION | | Authors: | Eron, S.J. | | Deposit date: | 2021-05-26 | | Release date: | 2021-11-03 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Development of an AchillesTAG degradation system and its application to control CAR-T activity
Curr Res Chem Biol, 1, 2021
|
|
7N03
 
 | | Crystal structure of MTH1 in complex with compound 31 | | Descriptor: | 4-anilino-6-(hexylamino)-N-methylquinoline-3-carboxamide, 7,8-dihydro-8-oxoguanine triphosphatase, SULFATE ION | | Authors: | Eron, S.J. | | Deposit date: | 2021-05-24 | | Release date: | 2021-11-03 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.13 Å) | | Cite: | Development of an AchillesTAG degradation system and its application to control CAR-T activity
Curr Res Chem Biol, 1, 2021
|
|
7NNJ
 
 | | Crystal Structure of NUDT4 (Diphosphoinositol polyphosphate phosphohydrolase 2) in complex with 4-O-Bn-1-PCP-InsP4 (AMR2105) | | Descriptor: | 1,2-ETHANEDIOL, Diphosphoinositol polyphosphate phosphohydrolase 2, FORMIC ACID, ... | | Authors: | Dubianok, Y, Arruda Bezerra, G, Raux, B, Diaz Saez, L, Riley, A.M, Potter, B.V.L, Huber, K.V.M, von Delft, F. | | Deposit date: | 2021-02-24 | | Release date: | 2022-03-23 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.755 Å) | | Cite: | Crystal Structure of NUDT4 (Diphosphoinositol polyphosphate phosphohydrolase 2) in complex with 4-O-Bn-1-PCP-InsP4 (AMR2105)
To Be Published
|
|
7NR6
 
 | | Structure of NUDT15 in complex with NSC56456 | | Descriptor: | 1,2-ETHANEDIOL, 2-azanyl-9-cyclohexyl-3~{H}-purine-6-thione, MAGNESIUM ION, ... | | Authors: | Rehling, D, Stenmark, P. | | Deposit date: | 2021-03-03 | | Release date: | 2022-03-23 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Coupling cellular target engagement to drug-induced responses with CeTEAM
To Be Published
|
|
4S2W
 
 | | Structure of E. coli RppH bound to sulfate ions | | Descriptor: | RNA pyrophosphohydrolase, SULFATE ION | | Authors: | Vasilyev, N, Serganov, A. | | Deposit date: | 2015-01-23 | | Release date: | 2015-02-11 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.992 Å) | | Cite: | Structures of RNA Complexes with the Escherichia coli RNA Pyrophosphohydrolase RppH Unveil the Basis for Specific 5'-End-dependent mRNA Decay.
J.Biol.Chem., 290, 2015
|
|
4S2X
 
 | | Structure of E. coli RppH bound to RNA and two magnesium ions | | Descriptor: | MAGNESIUM ION, RNA (5'-R(*(APC)*GP*U)-3'), RNA pyrophosphohydrolase, ... | | Authors: | Vasilyev, N, Serganov, A. | | Deposit date: | 2015-01-23 | | Release date: | 2015-02-11 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structures of RNA Complexes with the Escherichia coli RNA Pyrophosphohydrolase RppH Unveil the Basis for Specific 5'-End-dependent mRNA Decay.
J.Biol.Chem., 290, 2015
|
|
4S2Y
 
 | | Structure of E. coli RppH bound to RNA and three magnesium ions | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, RNA (5'-R(*(APC)*GP*U)-3'), ... | | Authors: | Vasilyev, N, Serganov, A. | | Deposit date: | 2015-01-23 | | Release date: | 2015-02-11 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structures of RNA Complexes with the Escherichia coli RNA Pyrophosphohydrolase RppH Unveil the Basis for Specific 5'-End-dependent mRNA Decay.
J.Biol.Chem., 290, 2015
|
|
4S2V
 
 | | E. coli RppH structure, KI soak | | Descriptor: | ACETATE ION, CALCIUM ION, IODIDE ION, ... | | Authors: | Vasilyev, N, Serganov, A. | | Deposit date: | 2015-01-23 | | Release date: | 2015-02-11 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structures of RNA Complexes with the Escherichia coli RNA Pyrophosphohydrolase RppH Unveil the Basis for Specific 5'-End-dependent mRNA Decay.
J.Biol.Chem., 290, 2015
|
|
7SP3
 
 | | E. coli RppH bound to Ap4A | | Descriptor: | BIS(ADENOSINE)-5'-TETRAPHOSPHATE, CHLORIDE ION, FLUORIDE ION, ... | | Authors: | Serganov, A.A, Vasilyev, N, Nuthanakanti, A. | | Deposit date: | 2021-11-02 | | Release date: | 2022-03-09 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | A distinct RNA recognition mechanism governs Np 4 decapping by RppH.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7SEZ
 
 | | Crystal structure of Vaccinia Virus decapping enzyme D9 in complex with m7GDP | | Descriptor: | 7N-METHYL-8-HYDROGUANOSINE-5'-DIPHOSPHATE, DNA repair NTP-phosphohydrolase, SODIUM ION | | Authors: | Peters, J.K, Tibble, R.W, Warminski, M, Jemielity, J, Gross, J.D. | | Deposit date: | 2021-10-02 | | Release date: | 2022-03-30 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.70001245 Å) | | Cite: | Structure of the poxvirus decapping enzyme D9 reveals its mechanism of cap recognition and catalysis.
Structure, 30, 2022
|
|
7SF0
 
 | | Crystal structure of Vaccinia Virus decapping enzyme D9 in complex with trinucleotide substrate | | Descriptor: | 7N-METHYL-8-HYDROGUANOSINE-5'-DIPHOSPHATE, DNA repair NTP-phosphohydrolase, MAGNESIUM ION, ... | | Authors: | Peters, J.K, Tibble, R.W, Warminski, M, Jemielity, J, Gross, J.D. | | Deposit date: | 2021-10-02 | | Release date: | 2022-03-30 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.95000446 Å) | | Cite: | Structure of the poxvirus decapping enzyme D9 reveals its mechanism of cap recognition and catalysis.
Structure, 30, 2022
|
|
3Q93
 
 | | Crystal Structure of Human 8-oxo-dGTPase (MTH1) | | Descriptor: | 7,8-dihydro-8-oxoguanine triphosphatase, GLYCEROL, IMIDAZOLE, ... | | Authors: | Tresaugues, L, Siponen, M.I, Arrowsmith, C.H, Berglund, H, Bountra, C, Collins, R, Edwards, A.M, Ekblad, T, Flodin, S, Flores, A, Graslund, S, Hammarstrom, M, Johansson, I, Karlberg, T, Kol, S, Kotenyova, T, Kouznetsova, E, Moche, M, Nyman, T, Persson, C, Schuler, H, Schutz, P, Thorsell, A.G, Van Der Berg, S, Wahlberg, E, Weigelt, J, Welin, M, Nordlund, P, Structural Genomics Consortium (SGC) | | Deposit date: | 2011-01-07 | | Release date: | 2011-03-02 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of Human 8-oxo-dGTPase (MTH1)
To be Published
|
|
3QSJ
 
 | | Crystal structure of NUDIX hydrolase from Alicyclobacillus acidocaldarius | | Descriptor: | CALCIUM ION, GLYCEROL, NUDIX hydrolase | | Authors: | Michalska, K, Wu, R, Bearden, J, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2011-02-21 | | Release date: | 2011-04-13 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of NUDIX hydrolase from Alicyclobacillus acidocaldarius
To be Published
|
|
7TN4
 
 | | Diphosphoinositol polyphosphate phosphohydrolase 1 (DIPP1/NUDT3) in complex with 3-diphosphoinositol 1,2,4,5-tetrakisphosphate (3-PP-IP4), Mg and Fluoride ion | | Descriptor: | (1R,2S,3R,4R,5S,6S)-4-hydroxy-2,3,5,6-tetrakis(phosphonooxy)cyclohexyl trihydrogen diphosphate, CHLORIDE ION, Diphosphoinositol polyphosphate phosphohydrolase 1, ... | | Authors: | Zong, G, Wang, H, Shears, S.B. | | Deposit date: | 2022-01-20 | | Release date: | 2022-06-15 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural and catalytic analyses of the InsP 6 kinase activities of higher plant ITPKs.
Faseb J., 36, 2022
|
|
7T7H
 
 | | Crystal structure of Vaccinia Virus decapping enzyme D9 in complex with inhibitor CP100356 | | Descriptor: | 4-(6,7-dimethoxy-3,4-dihydroisoquinolin-2(1H)-yl)-N-[2-(3,4-dimethoxyphenyl)ethyl]-6,7-dimethoxyquinazolin-2-amine, DNA repair NTP-phosphohydrolase, SODIUM ION | | Authors: | Peters, J.K, Gross, J.D. | | Deposit date: | 2021-12-15 | | Release date: | 2022-06-15 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.78000259 Å) | | Cite: | Fluorescence-Based Activity Screening Assay Reveals Small Molecule Inhibitors of Vaccinia Virus mRNA Decapping Enzyme D9.
Acs Chem.Biol., 17, 2022
|
|
3SHD
 
 | |
3SMD
 
 | |
