4LIJ
 
 | |
7CRE
 
 | |
8COO
 
 | | Solution structure of Zipcode binding protein 1 (ZBP1) KH3(DD)KH4 domains in complex with N6-Methyladenosine containing RNA | | Descriptor: | Insulin-like growth factor 2 mRNA-binding protein 1, RNA_(5'-R(*(UP*CP*GP*GP*(6MZ)P*CP*U)-3') | | Authors: | Nicastro, G, Abis, G, Taylor, I.A, Ramos, A. | | Deposit date: | 2023-02-28 | | Release date: | 2024-02-07 | | Method: | SOLUTION NMR | | Cite: | Direct m6A recognition by IMP1 underlays an alternative model of target selection for non-canonical methyl-readers.
Nucleic Acids Res., 51, 2023
|
|
3KRM
 
 | | Imp1 kh34 | | Descriptor: | GLYCEROL, Insulin-like growth factor 2 mRNA-binding protein 1 | | Authors: | Chao, J.A, Singer, R.H, Almo, S.C, Patskovsky, Y. | | Deposit date: | 2009-11-18 | | Release date: | 2010-02-09 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | ZBP1 recognition of beta-actin zipcode induces RNA looping.
Genes Dev., 24, 2010
|
|
1TUA
 
 | | 1.5 A Crystal Structure of a Protein of Unknown Function APE0754 from Aeropyrum pernix | | Descriptor: | Hypothetical protein APE0754 | | Authors: | Zhang, R, Skarina, T, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2004-06-24 | | Release date: | 2004-08-17 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | 1.5A crystal structure of a hypothetical protein APE0754 from Aeropyrum pernix
To be Published
|
|
4WAN
 
 | | Crystal structure of Msl5 protein in complex with RNA at 1.8 A | | Descriptor: | ACETATE ION, Branchpoint-bridging protein, GLYCEROL, ... | | Authors: | Jacewicz, A, Smith, P, Chico, L, Schwer, B, Shuman, S. | | Deposit date: | 2014-08-29 | | Release date: | 2014-12-17 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis for recognition of intron branchpoint RNA by yeast Msl5 and selective effects of interfacial mutations on splicing of yeast pre-mRNAs.
Rna, 21, 2015
|
|
4WAL
 
 | | Crystal structure of selenomethionine Msl5 protein in complex with RNA at 2.2 A | | Descriptor: | Branchpoint-bridging protein, CHLORIDE ION, GLYCEROL, ... | | Authors: | Jacewicz, A, Smith, P, Chico, L, Schwer, B, Shuman, S. | | Deposit date: | 2014-08-29 | | Release date: | 2014-12-17 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis for recognition of intron branchpoint RNA by yeast Msl5 and selective effects of interfacial mutations on splicing of yeast pre-mRNAs.
Rna, 21, 2015
|
|
6FBL
 
 | | NMR Solution Structure of MINA-1(254-334) | | Descriptor: | MINA-1 | | Authors: | Michel, E, Allain, F. | | Deposit date: | 2017-12-19 | | Release date: | 2019-01-30 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | MINA-1 and WAGO-4 are part of regulatory network coordinating germ cell death and RNAi in C. elegans.
Cell Death Differ., 26, 2019
|
|
6GQE
 
 | |
6QEY
 
 | | IMP1 KH1 and KH2 domains create a structural platform with unique RNA recognition and re-modelling properties | | Descriptor: | ACETONITRILE, Insulin-like growth factor 2 mRNA-binding protein 1, PHOSPHATE ION | | Authors: | Dagil, R, Ball, N.J, Ogrodowicz, R.W, Purkiss, A.G, Taylor, I.A, Ramos, A. | | Deposit date: | 2019-01-09 | | Release date: | 2019-03-27 | | Last modified: | 2019-09-18 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | IMP1 KH1 and KH2 domains create a structural platform with unique RNA recognition and re-modelling properties.
Nucleic Acids Res., 47, 2019
|
|
6Y2D
 
 | | Crystal structure of the second KH domain of FUBP1 | | Descriptor: | Far upstream element-binding protein 1, GLYCEROL, SULFATE ION | | Authors: | Ni, X, Chaikuad, A, Joerger, A.C, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-02-15 | | Release date: | 2020-03-25 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Comparative structural analyses and nucleotide-binding characterization of the four KH domains of FUBP1.
Sci Rep, 10, 2020
|
|
6Y2C
 
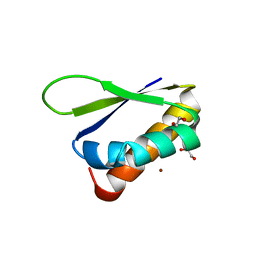 | | Crystal structure of the third KH domain of FUBP1 | | Descriptor: | 1,2-ETHANEDIOL, Far upstream element-binding protein 1, ZINC ION | | Authors: | Ni, X, Joerger, A.C, Chaikuad, A, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-02-15 | | Release date: | 2020-03-25 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Comparative structural analyses and nucleotide-binding characterization of the four KH domains of FUBP1.
Sci Rep, 10, 2020
|
|
4B8T
 
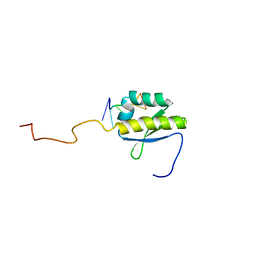 | | RNA BINDING PROTEIN Solution structure of the third KH domain of KSRP in complex with the G-rich target sequence. | | Descriptor: | 5'-R(*AP*GP*GP*GP*UP)-3', KH-TYPE SPLICING REGULATORY PROTEIN | | Authors: | Nicastro, G, Garcia-Mayoral, M.F, Hollingworth, D, Kelly, G, Martin, S.R, Briata, P, Gherzi, R, Ramos, A. | | Deposit date: | 2012-08-30 | | Release date: | 2012-11-07 | | Last modified: | 2013-01-16 | | Method: | SOLUTION NMR | | Cite: | Noncanonical G Recognition Mediates Ksrp Regulation of Let-7 Biogenesis
Nat.Struct.Mol.Biol., 19, 2012
|
|
6Y24
 
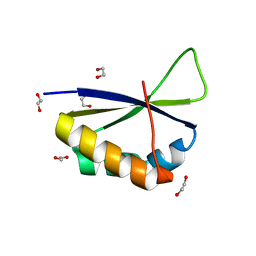 | | Crystal structure of fourth KH domain of FUBP1 | | Descriptor: | 1,2-ETHANEDIOL, Far upstream element-binding protein 1 | | Authors: | Ni, X, Joerger, A.C, Chaikuad, A, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-02-14 | | Release date: | 2020-03-25 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Comparative structural analyses and nucleotide-binding characterization of the four KH domains of FUBP1.
Sci Rep, 10, 2020
|
|
6ROL
 
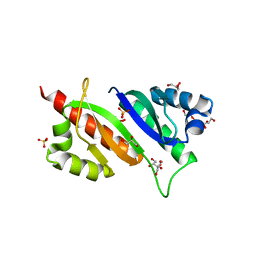 | | Structure of IMP2 KH34 domains | | Descriptor: | DI(HYDROXYETHYL)ETHER, GLYCEROL, Insulin-like growth factor 2 mRNA-binding protein 2, ... | | Authors: | Bhaskar, V, Biswas, J, Singer, R.H, Chao, J.A. | | Deposit date: | 2019-05-13 | | Release date: | 2019-10-16 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The structural basis for RNA selectivity by the IMP family of RNA-binding proteins.
Nat Commun, 10, 2019
|
|
3VKE
 
 | |
1J5K
 
 | |
1J4W
 
 | |
1K1G
 
 | | STRUCTURAL BASIS FOR RECOGNITION OF THE INTRON BRANCH SITE RNA BY SPLICING FACTOR 1 | | Descriptor: | 5'-R(*UP*AP*UP*AP*CP*UP*AP*AP*CP*AP*A)-3', SF1-Bo isoform | | Authors: | Liu, Z, Luyten, I, Bottomley, M.J, Messias, A.C, Houngninou-Molango, S, Sprangers, R, Zanier, K, Kramer, A, Sattler, M. | | Deposit date: | 2001-09-25 | | Release date: | 2001-11-07 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Structural basis for recognition of the intron branch site RNA by splicing factor 1.
Science, 294, 2001
|
|
1KHM
 
 | | C-TERMINAL KH DOMAIN OF HNRNP K (KH3) | | Descriptor: | PROTEIN (HNRNP K) | | Authors: | Baber, J, Libutti, D, Levens, D, Tjandra, N. | | Deposit date: | 1999-01-07 | | Release date: | 2000-01-12 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | High precision solution structure of the C-terminal KH domain of heterogeneous nuclear ribonucleoprotein K, a c-myc transcription factor.
J.Mol.Biol., 289, 1999
|
|
5WWW
 
 | | Crystal structure of the KH1 domain of human RNA-binding E3 ubiquitin-protein ligase MEX-3C complex with RNA | | Descriptor: | RNA (5'-R(*GP*UP*UP*UP*AP*G)-3'), RNA-binding E3 ubiquitin-protein ligase MEX3C | | Authors: | Yang, L, Wang, C, Li, F, Gong, Q. | | Deposit date: | 2017-01-05 | | Release date: | 2017-08-23 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.798 Å) | | Cite: | The human RNA-binding protein and E3 ligase MEX-3C binds the MEX-3-recognition element (MRE) motif with high affinity
J. Biol. Chem., 292, 2017
|
|
5WWZ
 
 | | Crystal structure of the KH2 domain of human RNA-binding E3 ubiquitin-protein ligase MEX-3C | | Descriptor: | RNA-binding E3 ubiquitin-protein ligase MEX3C, SULFATE ION | | Authors: | Yang, L, Wang, C, Li, F, Gong, Q. | | Deposit date: | 2017-01-05 | | Release date: | 2017-08-23 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The human RNA-binding protein and E3 ligase MEX-3C binds the MEX-3-recognition element (MRE) motif with high affinity
J. Biol. Chem., 292, 2017
|
|
5WWX
 
 | | Crystal structure of the KH2 domain of human RNA-binding E3 ubiquitin-protein ligase MEX-3C complex with RNA | | Descriptor: | NICKEL (II) ION, RNA (5'-R(P*AP*GP*AP*GP*U)-3'), RNA-binding E3 ubiquitin-protein ligase MEX3C | | Authors: | Yang, L, Wang, C, Li, F, Gong, Q. | | Deposit date: | 2017-01-05 | | Release date: | 2017-08-23 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The human RNA-binding protein and E3 ligase MEX-3C binds the MEX-3-recognition element (MRE) motif with high affinity
J. Biol. Chem., 292, 2017
|
|
1DT4
 
 | | CRYSTAL STRUCTURE OF NOVA-1 KH3 K-HOMOLOGY RNA-BINDING DOMAIN | | Descriptor: | NEURO-ONCOLOGICAL VENTRAL ANTIGEN 1 | | Authors: | Lewis, H.A, Chen, H, Edo, C, Buckanovich, R.J, Yang, Y.Y.L. | | Deposit date: | 2000-01-11 | | Release date: | 2000-02-11 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structures of Nova-1 and Nova-2 K-homology RNA-binding domains.
Structure Fold.Des., 7, 1999
|
|
1DTJ
 
 | | CRYSTAL STRUCTURE OF NOVA-2 KH3 K-HOMOLOGY RNA-BINDING DOMAIN | | Descriptor: | RNA-BINDING NEUROONCOLOGICAL VENTRAL ANTIGEN 2 | | Authors: | Lewis, H.A, Chen, H, Edo, C, Buckanovich, R.J, Yang, Y.Y.L, Musunuru, K, Zhong, R, Darnell, R.B, Burley, S.K. | | Deposit date: | 2000-01-12 | | Release date: | 2000-02-18 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structures of Nova-1 and Nova-2 K-homology RNA-binding domains.
Structure Fold.Des., 7, 1999
|
|
