5WSF
 
 | | Crystal structure of a cupin protein (tm1459) in osmium (Os)-substituted form II | | 分子名称: | OSMIUM ION, Uncharacterized protein tm1459 | | 著者 | Fujieda, N, Nakano, T, Taniguchi, Y, Ichihashi, H, Nishikawa, Y, Kurisu, G, Itoh, S. | | 登録日 | 2016-12-06 | | 公開日 | 2017-05-31 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.11 Å) | | 主引用文献 | A Well-Defined Osmium-Cupin Complex: Hyperstable Artificial Osmium Peroxygenase
J. Am. Chem. Soc., 2017
|
|
5WSE
 
 | | Crystal structure of a cupin protein (tm1459) in osmium (Os) substituted form I | | 分子名称: | OSMIUM ION, Uncharacterized protein tm1459 | | 著者 | Fujieda, N, Nakano, T, Taniguchi, Y, Ichihashi, H, Nishikawa, Y, Kurisu, G, Itoh, S. | | 登録日 | 2016-12-06 | | 公開日 | 2017-05-31 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.12 Å) | | 主引用文献 | A Well-Defined Osmium-Cupin Complex: Hyperstable Artificial Osmium Peroxygenase
J. Am. Chem. Soc., 2017
|
|
3LAG
 
 | | The crystal structure of a functionally unknown protein RPA4178 from Rhodopseudomonas palustris CGA009 | | 分子名称: | CALCIUM ION, FORMIC ACID, NICKEL (II) ION, ... | | 著者 | Tan, K, Evdokimova, E, Kudritska, M, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2010-01-06 | | 公開日 | 2010-01-19 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (1.15 Å) | | 主引用文献 | The crystal structure of a functionally unknown protein RPA4178 from Rhodopseudomonas palustris CGA009
To be Published
|
|
8HJX
 
 | |
8HJY
 
 | |
6L2D
 
 | | Crystal structure of a cupin protein (tm1459) in copper (Cu) substituted form | | 分子名称: | COPPER (II) ION, Cupin_2 domain-containing protein | | 著者 | Fujieda, N, Ichihashi, H, Nishikawa, Y, Kurisu, G, Itoh, S. | | 登録日 | 2019-10-03 | | 公開日 | 2020-04-01 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.198 Å) | | 主引用文献 | Cupin Variants as a Macromolecular Ligand Library for Stereoselective Michael Addition of Nitroalkanes.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
3HT1
 
 | |
5WSD
 
 | | Crystal structure of a cupin protein (tm1459) in apo form | | 分子名称: | Uncharacterized protein tm1459 | | 著者 | Fujieda, N, Nakano, T, Taniguchi, Y, Ichihashi, H, Nishikawa, Y, Kurisu, G, Itoh, S. | | 登録日 | 2016-12-06 | | 公開日 | 2017-05-31 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.2 Å) | | 主引用文献 | A Well-Defined Osmium-Cupin Complex: Hyperstable Artificial Osmium Peroxygenase
J. Am. Chem. Soc., 2017
|
|
6L2E
 
 | | Crystal structure of a cupin protein (tm1459, H52A mutant) in copper (Cu) substituted form | | 分子名称: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, COPPER (II) ION, Cupin_2 domain-containing protein | | 著者 | Fujieda, N, Ichihashi, H, Nishikawa, Y, Kurisu, G, Itoh, S. | | 登録日 | 2019-10-03 | | 公開日 | 2020-04-01 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.201 Å) | | 主引用文献 | Cupin Variants as a Macromolecular Ligand Library for Stereoselective Michael Addition of Nitroalkanes.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
8HJZ
 
 | |
6L2F
 
 | | Crystal structure of a cupin protein (tm1459, H54AH58A mutant) in copper (Cu) substituted form | | 分子名称: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 3,6,9,12,15,18-HEXAOXAICOSANE-1,20-DIOL, COPPER (II) ION, ... | | 著者 | Fujieda, N, Ichihashi, H, Nishikawa, Y, Kurisu, G, Itoh, S. | | 登録日 | 2019-10-03 | | 公開日 | 2020-04-01 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.23 Å) | | 主引用文献 | Cupin Variants as a Macromolecular Ligand Library for Stereoselective Michael Addition of Nitroalkanes.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
1V70
 
 | |
4Q29
 
 | | Ensemble Refinement of plu4264 protein from Photorhabdus luminescens | | 分子名称: | NICKEL (II) ION, SODIUM ION, plu4264 protein | | 著者 | Wang, F, Michalska, K, Li, H, Jedrzejczak, R, Babnigg, G, Bingman, C.A, Yennamalli, R, Weerth, S, Miller, M.D, Thomas, M.G, Joachimiak, A, Phillips Jr, G.N, Enzyme Discovery for Natural Product Biosynthesis (NatPro), Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2014-04-07 | | 公開日 | 2014-05-07 | | 最終更新日 | 2015-02-11 | | 実験手法 | X-RAY DIFFRACTION (1.349 Å) | | 主引用文献 | Structure of a cupin protein Plu4264 from Photorhabdus luminescens subsp. laumondii TTO1 at 1.35 angstrom resolution.
Proteins, 83, 2015
|
|
4MV2
 
 | | Crystal structure of plu4264 protein from Photorhabdus luminescens | | 分子名称: | NICKEL (II) ION, SODIUM ION, plu4264 | | 著者 | Michalska, K, Li, H, Jedrzejczak, R, Babnigg, G, Bingman, C.A, Yennamalli, R, Weerth, S, Thomas, M.G, Phillips Jr, G.N, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | 登録日 | 2013-09-23 | | 公開日 | 2013-10-02 | | 最終更新日 | 2015-02-04 | | 実験手法 | X-RAY DIFFRACTION (1.349 Å) | | 主引用文献 | Structure of a cupin protein Plu4264 from Photorhabdus luminescens subsp. laumondii TTO1 at 1.35 angstrom resolution.
Proteins, 83, 2015
|
|
5J4F
 
 | | Crystal structure of the N-terminally His6-tagged HP0902, an uncharacterized protein from Helicobacter pylori 26695 | | 分子名称: | Uncharacterized protein | | 著者 | Sim, D.-W, Lee, W.-C, Kim, H.Y, Kim, J.-H, Won, H.-S. | | 登録日 | 2016-04-01 | | 公開日 | 2017-02-08 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.4 Å) | | 主引用文献 | Structural identification of the lipopolysaccharide-binding capability of a cupin-family protein from Helicobacter pylori
FEBS Lett., 590, 2016
|
|
2GU9
 
 | |
2OA2
 
 | |
5TG0
 
 | |
2DCT
 
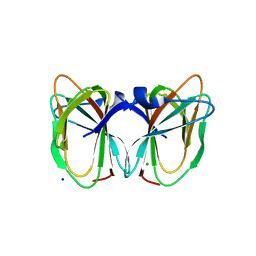 | | Crystal structure of the TT1209 from Thermus thermophilus HB8 | | 分子名称: | CHLORIDE ION, SODIUM ION, hypothetical protein TTHA0104 | | 著者 | Asada, Y, Sugahara, M, Shimizu, K, Yamamoto, H, Shimada, H, Nakamoto, T, Ono, N, Kunishima, N, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | 登録日 | 2006-01-12 | | 公開日 | 2006-01-24 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (1.45 Å) | | 主引用文献 | Crystal structure of the TT1209 from Thermus thermophilus HB8
To be Published
|
|
8AWN
 
 | | Crystal structure of a manganese-containing cupin (tm1459) from Thermotoga maritima, variant C106Q | | 分子名称: | CHLORIDE ION, Cupin_2 domain-containing protein | | 著者 | Grininger, C, Steiner, K, Gruber, K, Pavkov-Keller, T. | | 登録日 | 2022-08-30 | | 公開日 | 2023-03-08 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (1.45 Å) | | 主引用文献 | Engineering TM1459 for Stabilisation against Inactivation by Amino Acid Oxidation
Chem Ing Tech, 2023
|
|
7ZYB
 
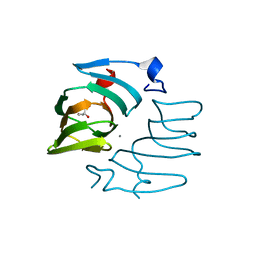 | |
5FPZ
 
 | | The structure of KdgF from Yersinia enterocolitica with malonate bound in the active site. | | 分子名称: | MALONIC ACID, NICKEL (II) ION, PECTIN DEGRADATION PROTEIN | | 著者 | Hobbs, J.K, Lee, S.M, Robb, M, Hof, F, Barr, C, Abe, K.T, Hehemann, J.H, McLean, R, Abbott, D.W, Boraston, A.B. | | 登録日 | 2015-12-03 | | 公開日 | 2016-05-04 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Kdgf, the Missing Link in the Microbial Metabolism of Uronate Sugars from Pectin and Alginate.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5FPX
 
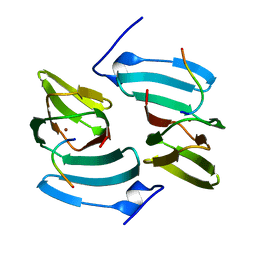 | | The structure of KdgF from Yersinia enterocolitica. | | 分子名称: | NICKEL (II) ION, PECTIN DEGRADATION PROTEIN, PEPTIDE | | 著者 | Hobbs, J.K, Lee, S.M, Robb, M, Hof, F, Barr, C, Abe, K.T, Hehemann, J.H, McLean, R, Abbott, D.W, Boraston, A.B. | | 登録日 | 2015-12-03 | | 公開日 | 2016-05-04 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Kdgf, the Missing Link in the Microbial Metabolism of Uronate Sugars from Pectin and Alginate.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
2O8Q
 
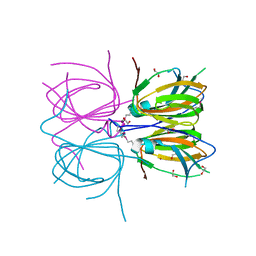 | |
4RD7
 
 | | The crystal structure of a Cupin 2 conserved barrel domain protein from Salinispora arenicola CNS-205 | | 分子名称: | Cupin 2 conserved barrel domain protein, GLYCEROL, SULFATE ION | | 著者 | Tan, K, Gu, M, Clancy, S, Phillips Jr, G.N, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | 登録日 | 2014-09-18 | | 公開日 | 2014-10-01 | | 最終更新日 | 2017-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.571 Å) | | 主引用文献 | The crystal structure of a Cupin 2 conserved barrel domain protein from Salinispora arenicola CNS-205
To be Published
|
|
